Summary: Study identifies a short gene segment crucial for brain development and information processing. The absence of the gene segment induces altered social behaviors, learning difficulties, and memory deficits, which are hallmarks of a subset of ASD.
Source: University of Toronto
Autism can bestow brilliance as well as cognitive difficulty, but how either scenario plays out in the brain is not clear. Now a study by University of Toronto researchers has found that a tiny gene fragment impacts the brain in a way that could explain swathes of autism cases that come with mental disability.
Researchers led by Benjamin Blencowe, a professor of molecular genetics in the Donnelly Centre for Cellular and Biomolecular Research, and Sabine Cordes, a senior investigator at Sinai Health System’s Lunenfeld-Tanenbaum Research Institute (LTRI), have identified a short gene segment that is crucial for brain development and information processing. Writing in the journal Molecular Cell, the researchers describe how an absence of this segment is sufficient to induce altered social behaviour–a hallmark of autism–in mice, as well as learning and memory deficits, which are seen in a subset of autism cases.
Best known for difficulties in social interaction and communication, autism is thought to arise from mishaps in brain wiring during development. It can strike in various ways–those who suffer from it can have superior mental ability or need full time care. Where on the autism spectrum a person falls depends in large part on their genetics, but most cases are idiopathic, or of unknown genetic origin.
“It’s very important to understand the mechanisms that underlie autism, especially in idiopathic forms where it is not clear what the underlying causes are,” says Thomas Gonatopoulos-Pournatzis, a research associate in Blencowe’s lab and lead author of the study. “Not only have we identified a new mechanism that contributes to this disorder, but our work may also offer a more rational development of therapeutic strategies.”
Blencowe’s team had previously uncovered a link between autism and short gene segments, known as microexons, that are predominantly expressed in the brain. Through a process known as alternative splicing, microexons are either spliced in or left out from the final gene transcript before it is translated into a protein. Although small, microexons can have dramatic effects by impacting a protein’s ability to bind its partners as required during brain development. However, how individual microexons contribute to autism is not clear.
The team focused on a specific microexon located in a gene known as eIF4G, which is critical for protein synthesis in the cell. They found that this microexon is overwhelmingly excluded from eIF4G gene transcripts in the brains of autistic individuals.
To test if the eIF4G microexon is important for brain function, Gonatopoulos-Pournatzis together with Cordes’ team bred mice that lack it. These mice showed social behaviour deficits, such as avoiding social interaction with other mice, establishing a link between the eIFG4 microexon and autistic-like behaviours.
A surprise came when the researchers found that these mice also performed poorly in a learning and memory test, which measures the animals’ ability to associate an environment with a stimulus.
“We could not have imagined that a single microexon would have such an important impact not only on social behaviour but also on learning and memory,” says Gonatopoulos-Pournatzis.
Further analysis revealed that the microexon encodes a part of eIF4G that allows it to associate with the Fragile X mental retardation protein, or FMRP, which is missing from people affected with Fragile X syndrome, a type of intellectual disability. About a third of individuals with Fragile X have features of autism but the link between the two remained unclear–until now.
eIF4G and FMRP associate in a complex that acts as a brake to hold off protein synthesis until new experience comes along, as the break is removed by neural activity, the researchers also found.
“It’s important to control brain responses to experience,” says Gonatopoulos-Pournatzis. “This brake in protein synthesis is removed upon experience and we think it allows formation of new memories.”
Without the microexon, however, this brake is weakened and what follows is increased protein production. The newly made proteins, identified in experiments performed with Anne-Claude Gingras, Senior Investigator at LTRI, form ion channels, receptors and other signaling molecules needed to build synapses and for them to function properly.

But, making too many of these proteins is not a good thing because this leads to the disruption of the type of brain waves involved in synaptic plasticity and memory formation, as revealed by electrode recordings of mouse brain slices, in experiments performed with the teams of Graham Collingridge, Senior Investigator at LTRI, and Melanie Woodin, a professor of cell and systems biology at U of T.
Moreover, an excess of similar kinds of proteins occurs in the absence of FMRP, suggesting a common molecular mechanism for Fragile X and idiopathic autism.
Researchers believe that their findings could help explain a substantial proportion of autism cases for which no other genetic clues are known. The findings also open the door to the development of new therapeutic approaches. One possibility is to increase the splicing of the eIF4G microexon in affected individuals using small molecules as a way to improve their social and cognitive deficits, Blencowe said.
The study would not have been possible without a close collaboration among multiple teams contributing diverse expertise. Blencowe and Gonatopoulos-Pournatzis also worked closely with Julie Forman-Kay, a professor of biochemistry and Program Head and Senior Scientist in the Molecular Medicine Program at the Hospital for Sick Children, and Nahum Sonenberg, a professor of biochemistry at McGill University.
Source:
University of Toronto
Media Contacts:
Jovana Drinjakovic – University of Toronto
Image Source:
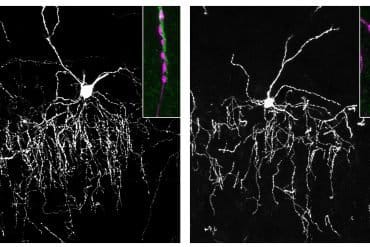
The image is credited to Thomas Gonatopoulos-Pournatzis.
Original Research: Closed access
“Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions”. Thomas Gonatopoulos-Pournatzis, Rieko Niibori, Eric W. Salter, Robert J. Weatheritt, Brian Tsang, Shaghayegh Farhangmehr, Xinyi Liang, Ulrich Braunschweig, and others.
Molecular Cell doi:10.1016/j.molcel.2020.01.006.
Abstract
Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions
Highlights
• Autism-disrupted eIF4G microexons mediate activity-dependent responses
• eIF4G microexons suppress the expression of critical synaptic proteins
• eIF4G microexon-deficient mice display social behavior and memory deficits
• eIF4G microexons coalesce with neuronal granule components and stall ribosomes
Summary
Microexons represent the most highly conserved class of alternative splicing, yet their functions are poorly understood. Here, we focus on closely related neuronal microexons overlapping prion-like domains in the translation initiation factors, eIF4G1 and eIF4G3, the splicing of which is activity dependent and frequently disrupted in autism. CRISPR-Cas9 deletion of these microexons selectively upregulates synaptic proteins that control neuronal activity and plasticity and further triggers a gene expression program mirroring that of activated neurons. Mice lacking the Eif4g1 microexon display social behavior, learning, and memory deficits, accompanied by altered hippocampal synaptic plasticity. We provide evidence that the eIF4G microexons function as a translational brake by causing ribosome stalling, through their propensity to promote the coalescence of cytoplasmic granule components associated with translation repression, including the fragile X mental retardation protein FMRP. The results thus reveal an autism-disrupted mechanism by which alternative splicing specializes neuronal translation to control higher order cognitive functioning.