Summary: A new study reveals how a bacterial species has adapted to the infant gut by producing LnbX, an enzyme that enables the microbe to grow on the sugar that is abundant only in human breast milk.
Source: Cell Press.
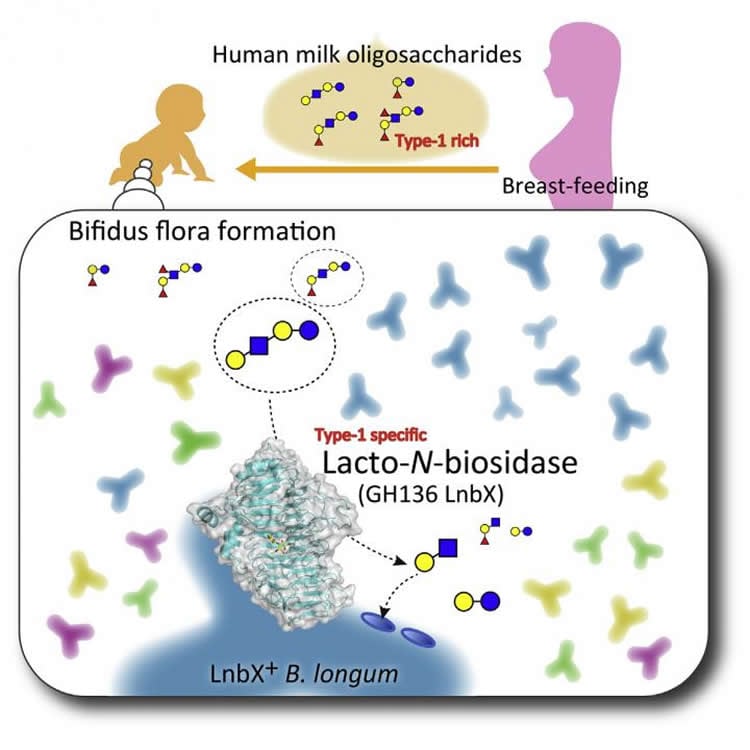
Breast milk provides vital nutrients not only to infants, but also to beneficial microbes that inhabit the gastrointestinal tract. A study published April 6 in Cell Chemical Biology shows that a bacterial species called Bifidobacterium longum has successfully adapted to the unique niche of the infant gut by producing an enzyme called LnbX, which enables this microbe to grow on a sugar that is abundant only in human milk.
“Given the health-promoting effects of bifidobacteria, our findings reveal a possible evolutionary route for the breast milk-driven symbiosis between gut microbes and humans,” says co-senior study author Takane Katayama of Kyoto University.
Gut microbes in early life are thought to have long-lasting effects on human health, and studies have shown that diet strongly influences the composition of this population. For example, human milk sugars are known to selectively promote the growth of beneficial gut microbes such as bifidobacteria, which prevent diarrhea and pathogenic infection in infants. One major component of human milk is a sugar called lacto-N-tetraose, which is virtually absent in the milk of other mammals. Bifidobacteria produce enzymes that break down this sugar, strongly suggesting that a symbiotic relationship recently evolved between these microorganisms and humans.
While investigating how this symbiotic relationship evolved, Katayama and co-senior study author Shinya Fushinobu of the University of Tokyo previously characterized LnbB and isolated LnbX — enzymes that degrade lacto-N-tetraose in Bifidobacterium bifidum and Bifidobacterium longum, respectively. In the new study, the researchers set out to build on these findings by determining the X-ray crystal structure of the catalytic domain of LnbX. The crystal structure, in combination with mutation and pharmacological experiments, revealed that LnbX has a distinct structure and catalytic mechanism from LnbB and therefore belongs to a novel family of glycoside hydrolase enzymes called GH136.
“Even though B. longum and B. bifidum belong to the same genus and inhabit the same environment, they use different enzymes to break down lacto-N-tetraose, taking advantage of the varied structures of this unique human milk sugar,” Fushinobu says. “The findings suggest that different strains and species of beneficial bifidobacteria have independently evolved distinct molecular tools to digest the same human milk sugar, explaining their ability to co-exist and thrive in the gastrointestinal tract of breast milk-fed infants.”
Additional experiments showed that the lnbX gene is critical for the ability of B. longum to grow specifically on lacto-N-tetraose. Moreover, an analysis of fecal DNA revealed that B. longum and the lnbX gene were more abundant in the gastrointestinal tract of ten infants that exclusively consumed breast milk compared with six infants fed a mixture of formula and breast milk. “Taken together, these findings suggest that lnbX is important for B. longum to persist in the gut ecosystem of breast milk-fed infants, and human milk sugars have been the main selective pressure for the evolution of lnbX,” Katayama says.
In future studies, the researchers will investigate whether other bifidobacterial species produce different enzymes to digest breast milk sugars. They will also look for bifidobacterial metabolites that promote infant health. In the end, this research could improve formula milk through fortification either with beneficial bacterial compounds, or with microbial enzymes that have been manipulated to carry out the reverse reaction and synthesize human milk sugars at a low cost and large scale.
“Although breast feeding during the first year of life is recommended in most cases, some mothers have to rely on formula milk because they have viral infections or do not produce milk with key nutrients such as zinc,” Katayama says. “Therefore, the development of new strategies to fortify formula milk with health-promoting ingredients will be especially critical in these rare cases where breast milk feeding is not possible.”
Funding: This work was supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, JSPS-KAKENHI, a Grant-in-Aid for JSPS Research Fellows, the Institution for Fermentation, Osaka, Japan, and in part by the Platform for Drug Discovery, Informatics, and Structural Life Science funded by MEXT.
Source: Joseph Caputo – Cell Press
Image Source: NeuroscienceNews.com image is credited to Yamada and Gotoh et al./Cell Chemical Biology 2017.
Original Research: Full open access research for “Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum” by Chihaya Yamada, Aina Gotoh, Mikiyasu Sakanaka, Mitchell Hattie, Keith A. Stubbs, Ayako Katayama-Ikegami, Junko Hirose, Shin Kurihara, Takatoshi Arakawa, Motomitsu Kitaoka, Shujiro Okuda, Takane Katayama, and Shinya Fushinobu in Cell Chemical Biology. Published online January 10 2017 doi:10.1016/j.chembiol.2017.03.012
[cbtabs][cbtab title=”MLA”]Cell Press “How a Beneficial Gut Microbe Adapted to Breast Milk.” NeuroscienceNews. NeuroscienceNews, 6 April 2017.
<https://neurosciencenews.com/lnbx-breast-milk-microbe-6353/>.[/cbtab][cbtab title=”APA”]Cell Press (2017, April 6). How a Beneficial Gut Microbe Adapted to Breast Milk. NeuroscienceNew. Retrieved April 6, 2017 from https://neurosciencenews.com/lnbx-breast-milk-microbe-6353/[/cbtab][cbtab title=”Chicago”]Cell Press “How a Beneficial Gut Microbe Adapted to Breast Milk.” https://neurosciencenews.com/lnbx-breast-milk-microbe-6353/ (accessed April 6, 2017).[/cbtab][/cbtabs]
Abstract
Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Highlights
•Crystal structure of a new lacto-N-biosidase (LnbX) from B. longum is presented
•The mechanism of LnbX is distinct from the lacto-N-biosidase of B. bifidum
•lnbX is indispensable for growth and occupancy in the breast-fed infant gut
•Host and microbe symbiotic and co-evolutionary relationships are revealed
Summary
Breast-fed infants generally have a bifidobacteria-rich microbiota with recent studies indicating that human milk oligosaccharides (HMOs) selectively promote bifidobacterial growth. Bifidobacterium bifidum possesses a glycoside hydrolase family 20 lacto-N-biosidase for liberating lacto-N-biose I from lacto-N-tetraose, an abundant HMO unique to human milk, while Bifidobacterium longum subsp. longum has a non-classified enzyme (LnbX). Here, we determined the crystal structure of the catalytic domain of LnbX and provide evidence for creation of a novel glycoside hydrolase family, GH136. The structure, in combination with inhibition and mutation studies, provides insight into the molecular mechanism and broader substrate specificity of this enzyme. Moreover, through genetic studies, we show that lnbX is indispensable for B. longum growth on lacto-N-tetraose and is a key genetic factor for persistence in the gut of breast-fed infants. Overall, this study reveals possible evolutionary routes for the emergence of symbiosis between humans and bifidobacterial species in the infant gut.
“Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum” by Chihaya Yamada, Aina Gotoh, Mikiyasu Sakanaka, Mitchell Hattie, Keith A. Stubbs, Ayako Katayama-Ikegami, Junko Hirose, Shin Kurihara, Takatoshi Arakawa, Motomitsu Kitaoka, Shujiro Okuda, Takane Katayama, and Shinya Fushinobu in Cell Chemical Biology. Published online January 10 2017 doi:10.1016/j.chembiol.2017.03.012