Summary: A new study claims researchers have proven multiple sclerosis can be caused by a single genetic mutation.
Source: University of British Columbia.
Scientists at the University of British Columbia and Vancouver Coastal Health have proven that multiple sclerosis (MS) can be caused by a single genetic mutation – a rare alteration in DNA that makes it very likely a person will develop the more devastating form of the neurological disease.
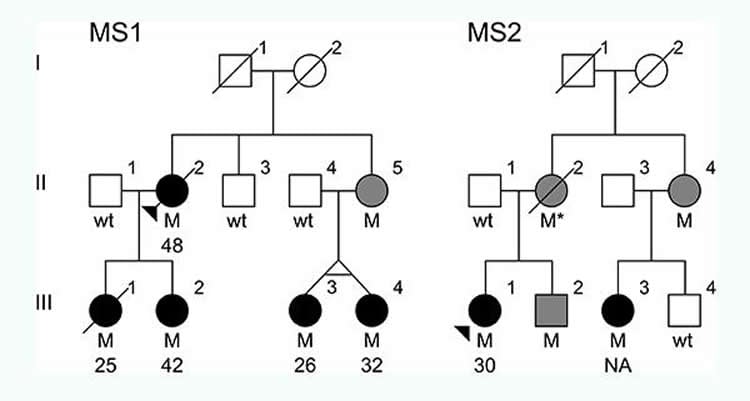
The mutation was found in two Canadian families that had several members diagnosed with a rapidly progressive type of MS, in which a person’s symptoms steadily worsen and for which there is no effective treatment.
The discovery of this mutation should erase doubts that at least some forms of MS are inherited. The prevailing view has been that a combination of many genetic variations cause a slight increase in susceptibility. In the two families described in this study, two-thirds of the people with the mutation developed the disease.
“This mutation puts these people at the edge of a cliff, but something still has to give them the push to set the disease process in motion,” said senior author Carles Vilarino-Guell, an assistant professor of medical genetics and a member of the Djavad Mowafaghian Centre for Brain Health.
Although only one in 1,000 MS patients appears to have this mutation, its discovery helps reveal the biological pathway that leads to the rapidly progressive form of the disease, accounting for about 15 per cent of people with MS. The discovery could also provide insight into the more common, fluctuating form of MS, known as “relapsing-remitting,” because in most cases, that disease gradually becomes progressive.
The findings, published today in the journal Neuron, could help in the search for therapies that act upon the gene itself or counteract the mutation’s disease-causing effects. More immediately, screening for the mutation in high-risk individuals could enable earlier diagnosis and treatment before symptoms appear.
“If you have this gene, chances are you will develop MS and rapidly deteriorate,” said co-author Dr. Anthony Traboulsee, the MS Society of Canada Research Chair at UBC and Director of Vancouver Coastal Health’s MS and Neuromyelitis Optica Clinic. “This could give us a critical early window of opportunity to throw everything at the disease, to try to stop it or slow it. Until now, we didn’t have much basis for doing that.”
Background on MS and this discovery
Signal disruption: Multiple sclerosis results from the body’s immune system attacking myelin, the fatty material that insulates neurons and enables rapid transmission of electrical signals. When myelin is damaged, communication between the brain and other parts of the body is disrupted, leading to vision problems, muscle weakness, difficulty with balance and coordination, and cognitive impairments. Canada has one of the highest rate of MS in the world, for reasons that elude scientists.
Defective protein: The MS-causing mutation discovered by UBC scientists was found on a gene called NR1H3. That gene produces a protein known as LXRA, which acts as an on-off switch on other genes. Some of those other genes stop the excessive inflammation that damages myelin or help create new myelin to repair the damage. A team led by Weihong Song, professor of psychiatry at UBC and the study’s other senior author, found that the mutation of that gene – a substitution of just one nucleotide for another in the DNA – produces a defective LXRA protein that is unable to activate those other critical genes.
Researchers at the University of British Columbia have discovered a rare genetic mutation that causes multiple sclerosis, which should erase doubts that some forms of the neurological disease are inherited.
Early intervention: People with a family history of MS could be screened for this mutation, and if they carry it, could be candidates for early diagnostic imaging long before symptoms appear; or they could opt to increase their intake of Vitamin D (low levels of which have been associated with the disease). If someone diagnosed with relapsing-remitting MS were found to have this mutation, they might be candidates for earlier, more aggressive treatment to delay the onset of the progressive form of the disease.
A more realistic animal model: Vilarino-Guell and his collaborators are now awaiting delivery of the first mice to be genetically engineered with this mutation, enabling examination of the cascade of reactions that leads to MS. Currently, scientists simulate MS in mice by either injecting them with myelin, which triggers an immune response, or by feeding them a drug that destroys myelin directly. Neither one mimics how the disease originates in humans.
Thousands of samples: The families with this mutation had donated to a Canadian-wide collection of blood samples from people with MS, begun in 1993 by co-author A. Dessa Sadovnick, a UBC professor of medical genetics and neurology. The 20-year project, funded by the MS Society of Canada and the Multiple Sclerosis Scientific Research Foundation, has samples from 4,400 people with MS, plus 8,600 blood relatives – one of the largest such biobanks in the world.
Funding: The study was funded by MS Society of Canada, Multiple Sclerosis Scientific Research Foundation.
Source: Brian Kladko – University of British Columbia
Image Source: This NeuroscienceNews.com image is credited to Carles Vilarino-Guell/University of British Columbia.
Video Source: The video is credited to UBC Medicine.
Original Research: Full open access research for “Nuclear Receptor NR1H3 in Familial Multiple Sclerosis” by Zhe Wang5, A. Dessa Sadovnick, Anthony L. Traboulsee, Jay P. Ross, Cecily Q. Bernales, Mary Encarnacion, Irene M. Yee, Madonna de Lemos, Talitha Greenwood, Joshua D. Lee, Galen Wright, Colin J. Ross, Si Zhang, Weihong Song, and Carles Vilariño-Güell in Neuron. Published online June 1 2016 doi:10.1016/j.neuron.2016.04.039
[cbtabs][cbtab title=”MLA”]University of British Columbia. “A Genetic Cause of Multiple Sclerosis Discovered.” NeuroscienceNews. NeuroscienceNews, 1 June 2016.
<https://neurosciencenews.com/multiple-sclerosis-genetics-4349/>.[/cbtab][cbtab title=”APA”]University of British Columbia. (2016, June 1). A Genetic Cause of Multiple Sclerosis Discovered. NeuroscienceNews. Retrieved June 1, 2016 from https://neurosciencenews.com/multiple-sclerosis-genetics-4349/[/cbtab][cbtab title=”Chicago”]University of British Columbia. “A Genetic Cause of Multiple Sclerosis Discovered.” https://neurosciencenews.com/multiple-sclerosis-genetics-4349/ (accessed June 1, 2016).[/cbtab][/cbtabs]
Abstract
Nuclear Receptor NR1H3 in Familial Multiple Sclerosis
Highlights
•An arginine to glutamine mutation in NR1H3 causes multiple sclerosis in families
•The clinical phenotype is consistent with rapidly progressive multiple sclerosis
•This mutation results in loss of function leading to transcriptional dysregulation
•Common variants in NR1H3 are associated with primary progressive multiple sclerosis
Summary
Multiple sclerosis (MS) is an inflammatory disease characterized by myelin loss and neuronal dysfunction. Despite the aggregation observed in some families, pathogenic mutations have remained elusive. In this study, we describe the identification of NR1H3 p.Arg415Gln in seven MS patients from two multi-incident families presenting severe and progressive disease, with an average age at onset of 34 years. Additionally, association analysis of common variants in NR1H3 identified rs2279238 conferring a 1.35-fold increased risk of developing progressive MS. The p.Arg415Gln position is highly conserved in orthologs and paralogs, and disrupts NR1H3 heterodimerization and transcriptional activation of target genes. Protein expression analysis revealed that mutant NR1H3 (LXRA) alters gene expression profiles, suggesting a disruption in transcriptional regulation as one of the mechanisms underlying MS pathogenesis. Our study indicates that pharmacological activation of LXRA or its targets may lead to effective treatments for the highly debilitating and currently untreatable progressive phase of MS.
“Nuclear Receptor NR1H3 in Familial Multiple Sclerosis” by Zhe Wang5, A. Dessa Sadovnick, Anthony L. Traboulsee, Jay P. Ross, Cecily Q. Bernales, Mary Encarnacion, Irene M. Yee, Madonna de Lemos, Talitha Greenwood, Joshua D. Lee, Galen Wright, Colin J. Ross, Si Zhang, Weihong Song, and Carles Vilariño-Güell in Neuron. Published online June 1 2016 doi:10.1016/j.neuron.2016.04.039