Collaboration led by Berkeley Lab researchers creates high-resolution map of gene regulatory elements in the brain.
Future research into the underlying causes of neurological disorders such as autism, epilepsy and schizophrenia, should greatly benefit from a first-of-its-kind atlas of gene-enhancers in the cerebrum (telencephalon). This new atlas, developed by a team led by researchers with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) is a publicly accessible Web-based collection of data that identifies and locates thousands of gene-regulating elements in a region of the brain that is of critical importance for cognition, motor functions and emotion.
“Understanding how the brain develops and functions, and how it malfunctions in neurological disorders, remains one of the most daunting challenges in contemporary science,” says Axel Visel, a geneticist with Berkeley Lab’s Genomics Division. “We’ve created a genome-wide digital atlas of gene enhancers in the human brain, the switches that tell genes when and where they need to be switched on or off. This enhancer atlas will enable other scientists to study in more detail how individual genes are regulated during development of the brain, and how genetic mutations may impact human neurological disorders.”
Visel is the corresponding author of a paper in the journal Cell that describes this work. The paper is titled “A High-Resolution Enhancer Atlas of the Developing Telencephalon.”

The cerebrum is the most highly developed region of the human brain. It houses the cerebral cortex, the so-called “gray matter” where complex information processing events take place, and the basal ganglia, a brain region that helps control movement throughout the body and is involved in certain types of learning. Many of the genes responsible for development of the cerebrum have been identified but most of the DNA elements responsible for expressing these genes, turning them on/off, have not. This is especially true for gene enhancers, sequences of DNA that act to amplify the expression of a specific gene. Characterizing gene enhancers tends to be difficult because an enhancer does not have to be located directly adjacent to the gene it is enhancing, but can in fact be located hundreds of thousands of DNA basepairs away.
“In addition to acting over long distances and being located upstream, downstream or in introns of protein-coding genes, the sequence features of gene enhancers are poorly understood,” Visel says. “However, gene-centric studies have provided strong evidence that gene enhancers are critical for normal embryonic development of the brain and have also linked human diseases to perturbed enhancer sequences.”
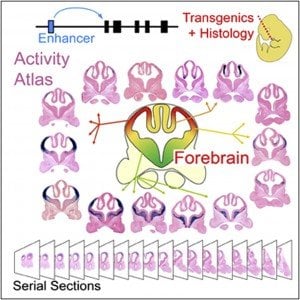
Visel and an international team of researchers met the challenges of systematic identification and functional characterization of gene enhancers in the cerebrum through a combination of ChIP-seq studies and large-scale histological analyses in transgenic mice, in which the activity patterns of human telencephalon enhancers can be studied. ChIP-seq, which stands for “chromatin immunoprecipitation followed by sequencing,” is a technique for genome-wide profiling of proteins that interact with DNA.
This combination of approaches enabled Visel and his colleagues to identify over 4,600 candidate embryonic forebrain enhancers. Furthermore, studying mouse embryos they mapped the activity of 145 of these enhancers at high resolution to define where exactly in the developing brain they drive the expression of their respective target genes. The result is a comprehensive, electronically accessible database for investigating the gene regulatory mechanisms of cerebrum development, and for studying the roles of distant-acting enhancers in neurodevelopmental disorders.

“By mapping hundreds of gene enhancer sequences and defining where exactly in the developing brain they are active, our enhancer atlas provides important information to connect non-coding mutations to actual biological functions,” Visel says.
As an example of how the enhancer atlas could be used in this manner, Visel says that if through genetic studies a certain region of the human genome has been linked to a specific neurological disorder but that region does not contain any protein-coding genes, checking the atlas could reveal whether the region is home to any distant-acting gene enhancers.
“If there is a gene enhancer in the region,” Visel says, “researchers could determine exactly how the enhancer or a mutated form is related to the disorder. This encompasses a lot of territory in the human genome. More than half of all the disease-related DNA sequence changes discovered in genome-wide screens to date fall within the 98-percent of human genome sequences that do not code for proteins. These ‘non-coding’ sequences were once dismissed by many as ‘junk DNA’ but are now known to harbor critical gene regulatory functions.”
Visel says the enhancer atlas also provides a “molecular toolbox” that can be used to re-locate the expression of certain genes to specific regions of the brain in experimental applications. This could help researchers gain new insights into how the brain develops and how the human brain has evolved.
“The evolution of the gene regulatory architecture may have been one of the drivers of the extremely advanced brains that we see in primates in general and humans in particular,” he says.
While continuing to map and annotate additional enhancers, Visel and his colleagues are now also studying the biological mechanisms related to some of these enhancers in more detail.
“We still only partially understand how gene enhancers work in the context of a living brain, i.e., what proteins bind to them, how mutations change their function, and what exactly the role of these sequences is in the context of human neurological disorders,” Visel says. “Clearly, there is more work to do.”
Notes about this genetics and neuroscience research article
Co-authoring the Cell paper with Visel were Leila Taher, Hani Girgis, Dalit May, Olga Golonzhka, Renee Hoch, Gabriel McKinsey, Kartik Pattabiraman, Shanni Silberberg, Matthew Blow, David Hansen, Alex S. Nord, Jennifer Akiyama, Amy Holt, Roya Hosseini, Sengthavy Phouanenavong, Ingrid Plajzer-Frick, Malak Shoukry, Veena Afzal, Tommy Kaplan, Arnold Kriegstein, Edward Rubin, Ivan Ovcharenko, Len Pennacchio and John Rubenstein.
This research was primarily supported by the National Institutes of Health. Studies were performed at all of the collaborating institutions.
Contact: Lynn Yarris – Berkeley Lab
Source: Berkeley Lab press release
Image Source: All images are adapted from press release by Berkeley Labs.
Original Research: Abstract for “A High-Resolution Enhancer Atlas of the Developing Telencephalon” by Axel Visel, Leila Taher, Hani Girgis, Dalit May, Olga Golonzhka, Renee V. Hoch, Gabriel L. McKinsey, Kartik Pattabiraman, Shanni N. Silberberg, Matthew J. Blow, David V. Hansen, Alex S. Nord, Jennifer A. Akiyama, Amy Holt, Roya Hosseini, Sengthavy Phouanenavong, Ingrid Plajzer-Frick, Malak Shoukry, Veena Afzal, Tommy Kaplan, Arnold R. Kriegstein, Edward M. Rubin, Ivan Ovcharenko, Len A. Pennacchio, John L.R. Rubenstein in Cell. Published online January 31 2013 http://dx.doi.org/10.1016/j.cell.2012.12.041