Further underscoring the prenatal origins of Autism Spectrum Disorder (ASD), researchers at University of California, San Diego School of Medicine describe for the first time how abnormal gene activity in cell cycle networks that are known to control brain cell production may underlie abnormal early brain growth in the disorder.
The findings are published online December 14 in Molecular Systems Biology.
“These findings identify common genomic defects that help explain why there are abnormal numbers of brain cells in autism, why the brain grows abnormally too large or too small in some ASD toddlers and how previously reported diverse gene mutations may, in fact, converge in their effects via common genomic pathways,” said senior study author Eric Courchesne, PhD, professor of neurosciences and director of the Autism Center of Excellence at UC San Diego.
“This study, along with the work of others and our past findings on excess neurons and brain growth patterns, enable researchers to now trace the origins of early abnormal brain overgrowth in ASD back to prenatal disruption of networks governing brain cell numbers.”
The proper regulation of brain cell numbers is one of earliest critical steps in fetal brain development. Courchesne and colleagues first discovered in 2001 that early brain overgrowth occurs in a substantial subset of ASD toddlers. In 2011, they reported that the young ASD prefrontal cortex, the region of the brain associated with social and communications skills, exhibited a 67 percent excess in cortical cells. In 2014, they described patches of disrupted cortical development in the brains of ASD children.
Rare gene mutations capable of increasing or decreasing cell number and brain size have been reported for tiny percentages of all ASD individuals, but for the vast majority of ASD children, the genomic defects behind abnormal brain overgrowth or undergrowth have remained unknown. The new study points to a common underlying defective functional genomic network – cell cycle – in living ASD toddlers, which plays a central role in fetal brain development.
Courchesne noted that several hundred different genes coordinate the cell cycle network in the fetal brain. “When they are active in the right way at the right time, they generate the correct number and type of brain cells that go to the cortex to create the normal brain cell layers and connections,” he said. “Both genetic mutations and non-genetic factors can change how these genes work and cause abnormal cell numbers and cell types.”
Attempts to better understand the genetic mechanisms underlying ASD are hindered by, among other things, the impossibility of direct brain gene expression measurement during critical periods of early development. However, first author Tiziano Pramparo, PhD, said blood and brain tissues highly express many of the same gene networks, particularly those related to cell cycle functions.
In the current study, researchers investigated the relationships between blood gene expression and brain size in 142 male toddlers, ages 1 to 4 years, 87 with a diagnosis of ASD and 55 controls.
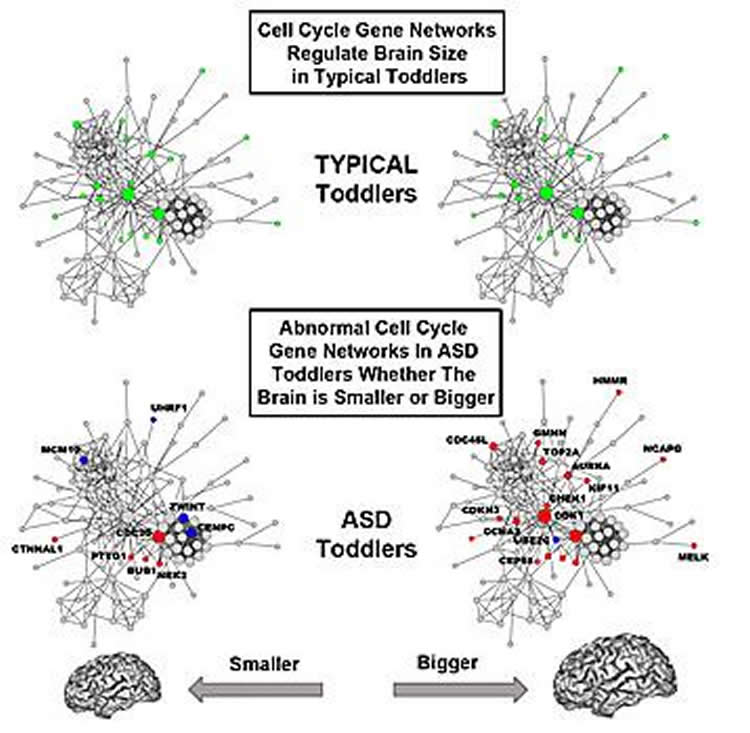
In the control toddlers, researchers found that variation in brain size significantly correlated with cell cycle and protein folding gene networks, potentially impacting neuron number and synapse development. In ASD toddlers, correlations with brain size were strikingly disrupted due to considerable changes in the organization of cell cycle gene networks. Conversely, cell adhesion gene networks, which regulate the ability of cells to stick together to form multicellular tissues and communicate, showed an abnormal relationship with variation in brain size in ASD toddlers.
They also identified 23 candidate genes for brain maldevelopment, of which 4 are directly linked to genes frequently mutated in ASD. “These patterns of activity in these 23 genes relative to brain size in the ASD toddlers was very different from that in typical toddlers,” observed Pramparo.
Pramparo said the new findings provide in vivo evidence for the involvement of cell cycle processes in ASD brain maldevelopment and significantly illuminate the complexities involved in early dysregulation and disruption of the developing ASD brain. Specifically, he noted:
- They point to a convergence of findings related to mutation, gene expression and brain-related phenotypes in ASD, “which is key since ASD is very heterogeneous, both at the genetic and clinical level.”
- Many “high-confidence” ASD candidate genes are upstream and may control expression of other genes and networks with primary cell cycle functions.
- Timing of the cell cycle appears to be critical. Shorter cell cycle periods may result in bigger brains and greater dysregulation. “The difference in smaller versus bigger may be due to genetic backgrounds, but also to non-genetic triggering events, especially when they occur during development.”
Courchesne said the findings raise the possibility that future research could test whether genomic biomarkers of cell cycle disruption could be an early sign of ASD risk or might be indicators of clinical progression, severity and outcome.
“Our next studies will examine whether infants at risk for ASD who have the most deviant cell cycle expression patterns also have more abnormal early development of brain structure, connections and function,” he said. “The findings do not directly suggest new therapies for children with ASD, but they raise the important question of whether the degree of genomic disruption might be related to treatment responsiveness.”
Exploring this question, he said, along with how development of neural connectivity and functional patterns is altered in ASD brains, will be addressed in future research.
Co-authors include Kathleen Campbell, Cynthia Carter Barnes, Steven Marinero, Stephanie Solso, Julia Young, Maisi Mayo, Anders Dale, Clelia Ahrens-Barbeau, Linda Lopez, Nathan Lewis and Karen Pierce, UC San Diego; Michael V. Lombardo, University of Cyprus and University of Cambridge; and Sarah S. Murray, The Scripps Translational Sciences Institute and UC San Diego.
Funding: Funding for this research came, in part, from the National Institutes of Health (grants P50-MH081755, R01-MH0800134, R01-MH036840, 1U54RR025205-0); the UC San Diego Clinical and Translational Research Institute (grants KL2TR00099 AND 1KL2TR001444) and the Novo Nordisk Foundation.
Source: Scott LaFee – UCSD
Image Credit: The image is credited to UC San Diego Health
Original Research: Full open access research for “Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers” by Tiziano Pramparo, Michael V Lombardo, Kathleen Campbell, Cynthia Carter Barnes, Steven Marinero, Stephanie Solso, Julia Young, Maisi Mayo, Anders Dale, Clelia Ahrens‐Barbeau, Sarah S Murray, Linda Lopez, Nathan Lewis, Karen Pierce, and Eric Courchesne in Molecular Systems Biology. Published online December 14 2015 doi:10.15252/msb.20156108
Abstract
Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers
Genetic mechanisms underlying abnormal early neural development in toddlers with Autism Spectrum Disorder (ASD) remain uncertain due to the impossibility of direct brain gene expression measurement during critical periods of early development. Recent findings from a multi‐tissue study demonstrated high expression of many of the same gene networks between blood and brain tissues, in particular with cell cycle functions. We explored relationships between blood gene expression and total brain volume (TBV) in 142 ASD and control male toddlers. In control toddlers, TBV variation significantly correlated with cell cycle and protein folding gene networks, potentially impacting neuron number and synapse development. In ASD toddlers, their correlations with brain size were lost as a result of considerable changes in network organization, while cell adhesion gene networks significantly correlated with TBV variation. Cell cycle networks detected in blood are highly preserved in the human brain and are upregulated during prenatal states of development. Overall, alterations were more pronounced in bigger brains. We identified 23 candidate genes for brain maldevelopment linked to 32 genes frequently mutated in ASD. The integrated network includes genes that are dysregulated in leukocyte and/or postmortem brain tissue of ASD subjects and belong to signaling pathways regulating cell cycle G1/S and G2/M phase transition. Finally, analyses of the CHD8 subnetwork and altered transcript levels from an independent study of CHD8 suppression further confirmed the central role of genes regulating neurogenesis and cell adhesion processes in ASD brain maldevelopment.
“Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers” by Tiziano Pramparo, Michael V Lombardo, Kathleen Campbell, Cynthia Carter Barnes, Steven Marinero, Stephanie Solso, Julia Young, Maisi Mayo, Anders Dale, Clelia Ahrens‐Barbeau, Sarah S Murray, Linda Lopez, Nathan Lewis, Karen Pierce, and Eric Courchesne in Molecular Systems Biology. Published online December 14 2015 doi:10.15252/msb.20156108