Summary: Microscopy technology allows researchers to discover new connections in brain areas associated with memory and learning in fruit flies.
Source: HHMI.
What was once thought to be a done-and-dusted map of the fruit fly brain has gotten a second look, and researchers have discovered that it’s actually not done at all.
Two teams of scientists at the Janelia Research Campus have independently mapped a brain region critical for memory and learning in the fruit fly, Drosophila melanogaster, in the larval and adult life stages. Their findings have added some unexpected neural connections, or synapses, to a seemingly solved circuit.
The results underscore the need for deeply detailed brain mapping, and make a case for using high-resolution electron microscopy (EM) to do it, says Albert Cardona, a group leader and neurobiologist at Janelia. Even in the most well-studied parts of the fly brain, he says, there’s still a lot more for scientists to see.
Cardona and colleagues, and Janelia’s FlyEM Project team and collaborators, recently published two separate studies: one in Nature on August 10, 2017, the other in eLife on July 18, 2017, detailing the newly discovered neural connections and how they might relate to behavior.
“This region of the brain had been picked over for decades, and people had already described the major circuits,” says Gerry Rubin, Janelia’s executive director. “The models had everything all neat and tidy; these papers say, ‘Well, not so quick.’ ”
Scientists have studied the neural underpinnings of fly learning and memory for more than 40 years, and mapped the brain region involved using light microscopy. That technique doesn’t capture the neural nitty-gritty in quite the same way that electron microscopy does, Rubin says. EM is more time-intensive but can offer much more imaging detail.
Rubin likens the current state of EM in connectomics, the mapping of synapses in the brain, to the early days of human genome sequencing. Rubin, a pioneer of gene sequencing in Drosophila during his tenure as an HHMI investigator at the University of California, Berkeley, recalls that some scientists initially shied away from sequencing. They cited the deluge of information and deep detail as a burden, not a boon, he says. Now a similar dispute permeates the connectomics field: Is electron microscopy worth the additional effort? Or can scientists continue to rely on tried-and-true light microscopy?
Now, given Cardona and FlyEM’s new studies, the teams say that EM should be included as a key aspect of understanding the fly brain.
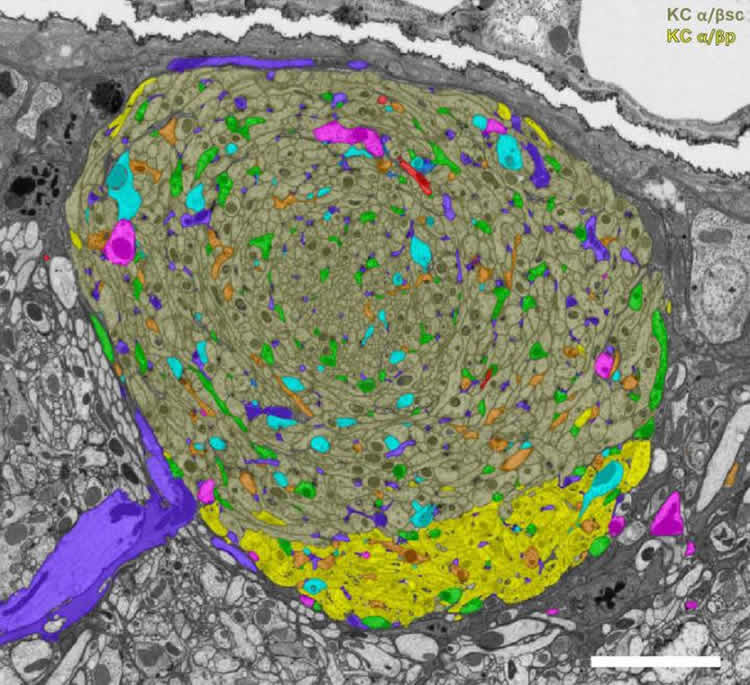
Both teams’ papers are part of larger efforts to map the whole Drosophila nervous system – Cardona and collaborators in larvae, the FlyEM Project’s team in adult flies. The studies employed two different EM methods: transmission electron microscopy (TEM) in the larvae and focused-ion beam scanning electron microscopy (FIB-SEM) in the adult, and specialized software that helps make sense of the immense tangle of neurons. Both studies targeted the mushroom body, the area of the brain responsible for associative learning (like associating a bell’s ring with a tasty treat – think Pavlov’s dog).
The mushroom body consists of multiple compartments, three of which make up the “alpha lobe” in the adult. FlyEM zeroed in on this region, mapping a total of 983 neurons. Cardona’s approach differed slightly. Neurons in the larva’s nervous system are smaller compared to the adult fly, so he was able to trace the 300 neurons of the larval mushroom bodies.
Much to their surprise, FIB-SEM revealed three classes of never-before-seen synapses in the mushroom body – which were observed in both the larva and the adult. These synaptic connections were formed among three neuron types: Kenyon cells, which report sensory information; dopaminergic neurons, which provide details about the sensory information; and output neurons, which relay information beyond the mushroom body.
The fact that these synapses were seen in young and full-grown flies implies that the connections are a critical part of the learning and memory circuit, Rubin says.
“When we saw them in the larva, we weren’t sure if it was just a developmental artifact – if they would go away, like the neuron version of baby teeth,” he says. Janelia’s FlyEM team confirmed the neurons’ existence in adults, and study coauthors, Yoshi Aso, Toshi Hige, and Glenn Turner, showed that the synapses contributed to the function of the mushroom body.
Now, the teams are continuing to map the fly and larva nervous systems in full. FlyEM, with 10-fold more neurons to chart, is taking advantage of a boost from the software side – applying the latest machine vision and artificial intelligence methods in collaboration with Google, as well as computer algorithms and software developed in-house at Janelia. Steve Plaza, who manages the FlyEM team, speculates that the 18 months it took to complete this study could have stretched to 10 years had it not been for the computational power that streamlined their work.
“I’m really excited about the progress in this field,” Rubin says. “If you would have asked me a year ago how long it would take to map the whole fly brain, I would have said, ‘Thirty years, and I’d be lucky to see it.’ Now, I’d say more like five years,” he says. “I think the field is really about to take off.”
Source: Meghan Rosen – HHMI
Publisher: Organized by NeuroscienceNews.com.
Image Source: NeuroscienceNews.com images are credited to Takemura et al./ eLife 2017.
Video Source: Video credited to HHMI Howard Hughes Medical Institute.
Original Research: Full open access research for “A connectome of a learning and memory center in the adult Drosophila brain” by Shin-ya Takemura, Yoshinori Aso, Toshihide Hige, Allan Wong, Zhiyuan Lu, C. Shan Xu, Patricia K. Rivlin, Harald Hess, Ting Zhao, Toufiq Parag, Stuart Berg, Gary Huang, William Katz, Donald J. Olbris, Stephen Plaza, Lowell Umayam, Roxanne Aniceto, Lei-Ann Chang, Shirley Lauchie, Omotara Ogundeyi, Christopher Ordish, Aya Shinomiya, Christopher Sigmund, Satoko Takemura, Julie Tran, Glenn C. Turner, Gerald M. Rubin, and Louis K. Scheffer in eLife. Published online July 18 2017 doi:10.7554/eLife.26975
Abstract for “The complete connectome of a learning and memory centre in an insect brain” by Katharina Eichler, Feng Li, Ashok Litwin-Kumar, Youngser Park, Ingrid Andrade, Casey M. Schneider-Mizell, Timo Saumweber, Annina Huser, Claire Eschbach, Bertram Gerber, Richard D. Fetter, James W. Truman, Carey E. Priebe, L. F. Abbott, Andreas S. Thum, Marta Zlatic & Albert Cardona in Nature. Published online August 9 2017 doi:10.1038/nature23455
[cbtabs][cbtab title=”MLA”]HHMI “Electron Microscopy Uncovers Unexpected Connections in Fruit Fly Brain.” NeuroscienceNews. NeuroscienceNews, 3 November 2017.
<https://neurosciencenews.com/fruit-fly-brain-connection-7864/>.[/cbtab][cbtab title=”APA”]HHMI (2017, November 3). Electron Microscopy Uncovers Unexpected Connections in Fruit Fly Brain. NeuroscienceNews. Retrieved November 3, 2017 from https://neurosciencenews.com/fruit-fly-brain-connection-7864/[/cbtab][cbtab title=”Chicago”]HHMI “Electron Microscopy Uncovers Unexpected Connections in Fruit Fly Brain.” https://neurosciencenews.com/fruit-fly-brain-connection-7864/ (accessed November 3, 2017).[/cbtab][/cbtabs]
Abstract
A connectome of a learning and memory center in the adult Drosophila brain
Understanding memory formation, storage and retrieval requires knowledge of the underlying neuronal circuits. In Drosophila, the mushroom body (MB) is the major site of associative learning. We reconstructed the morphologies and synaptic connections of all 983 neurons within the three functional units, or compartments, that compose the adult MB’s α lobe, using a dataset of isotropic 8 nm voxels collected by focused ion-beam milling scanning electron microscopy. We found that Kenyon cells (KCs), whose sparse activity encodes sensory information, each make multiple en passant synapses to MB output neurons (MBONs) in each compartment. Some MBONs have inputs from all KCs, while others differentially sample sensory modalities. Only 6% of KC>MBON synapses receive a direct synapse from a dopaminergic neuron (DAN). We identified two unanticipated classes of synapses, KC>DAN and DAN>MBON. DAN activation produces a slow depolarization of the MBON in these DAN>MBON synapses and can weaken memory recall.
“A connectome of a learning and memory center in the adult Drosophila brain” by Shin-ya Takemura, Yoshinori Aso, Toshihide Hige, Allan Wong, Zhiyuan Lu, C. Shan Xu, Patricia K. Rivlin, Harald Hess, Ting Zhao, Toufiq Parag, Stuart Berg, Gary Huang, William Katz, Donald J. Olbris, Stephen Plaza, Lowell Umayam, Roxanne Aniceto, Lei-Ann Chang, Shirley Lauchie, Omotara Ogundeyi, Christopher Ordish, Aya Shinomiya, Christopher Sigmund, Satoko Takemura, Julie Tran, Glenn C. Turner, Gerald M. Rubin, and Louis K. Scheffer in eLife. Published online July 18 2017 doi:10.7554/eLife.26975
Abstract
The complete connectome of a learning and memory centre in an insect brain
Associating stimuli with positive or negative reinforcement is essential for survival, but a complete wiring diagram of a higher-order circuit supporting associative memory has not been previously available. Here we reconstruct one such circuit at synaptic resolution, the Drosophila larval mushroom body. We find that most Kenyon cells integrate random combinations of inputs but that a subset receives stereotyped inputs from single projection neurons. This organization maximizes performance of a model output neuron on a stimulus discrimination task. We also report a novel canonical circuit in each mushroom body compartment with previously unidentified connections: reciprocal Kenyon cell to modulatory neuron connections, modulatory neuron to output neuron connections, and a surprisingly high number of recurrent connections between Kenyon cells. Stereotyped connections found between output neurons could enhance the selection of learned behaviours. The complete circuit map of the mushroom body should guide future functional studies of this learning and memory centre.
“The complete connectome of a learning and memory centre in an insect brain” by Katharina Eichler, Feng Li, Ashok Litwin-Kumar, Youngser Park, Ingrid Andrade, Casey M. Schneider-Mizell, Timo Saumweber, Annina Huser, Claire Eschbach, Bertram Gerber, Richard D. Fetter, James W. Truman, Carey E. Priebe, L. F. Abbott, Andreas S. Thum, Marta Zlatic & Albert Cardona in Nature. Published online August 9 2017 doi:10.1038/nature23455






