Summary: Study reveals how different signals in the brain are coded in the vagus nerve, finding three key features that are independently coded by vagal sensory neurons.
Source: Yale
The human brain is a busy organ—detecting signals from all over the body as it undergoes change throughout the day. When the lungs inhale an irritant, the body knows to cough. Or when the stomach ingests toxins, it induces vomiting. The brain plays a role in both.
The brain’s ability to precisely discriminate among various signals has fascinated scientists, but the biological mechanism is still unclear. Now, in a new study that aims to understand how different signals in the body are coded in the vagus nerve—the cranial nerve that sends information to and from the brain about internal organ function—Yale researchers have found that the signals have three key features that are independently coded by vagal sensory neurons.
They are: which organ a signal is coming from, which tissue layer within the organ the signal is coming from, and what the stimulus is. This coding enables the high precision achieved by the brain.
The researchers, including co-senior authors Rui Chang, Ph.D., assistant professor of neuroscience and of cellular & molecular physiology, and Le Zhang, Ph.D., assistant professor of neurology, published their study in Nature on March 16.
Tracing intricate links between brain and body
The body’s ability to sense changes within itself is called interoception, a process that is essential to survival. This body-to-brain connection is made through the vagus nerve, and the signals received by that nerve are coded independently by specialized vagal sensory neurons.
“This is the first time we actually know how different body signals are being represented through the vagal interoception system to the brain in a very precise and accurate manner,” says Chang. “We know that the brain can very precisely discriminate signals, but what is the biological reason for that discrimination?”
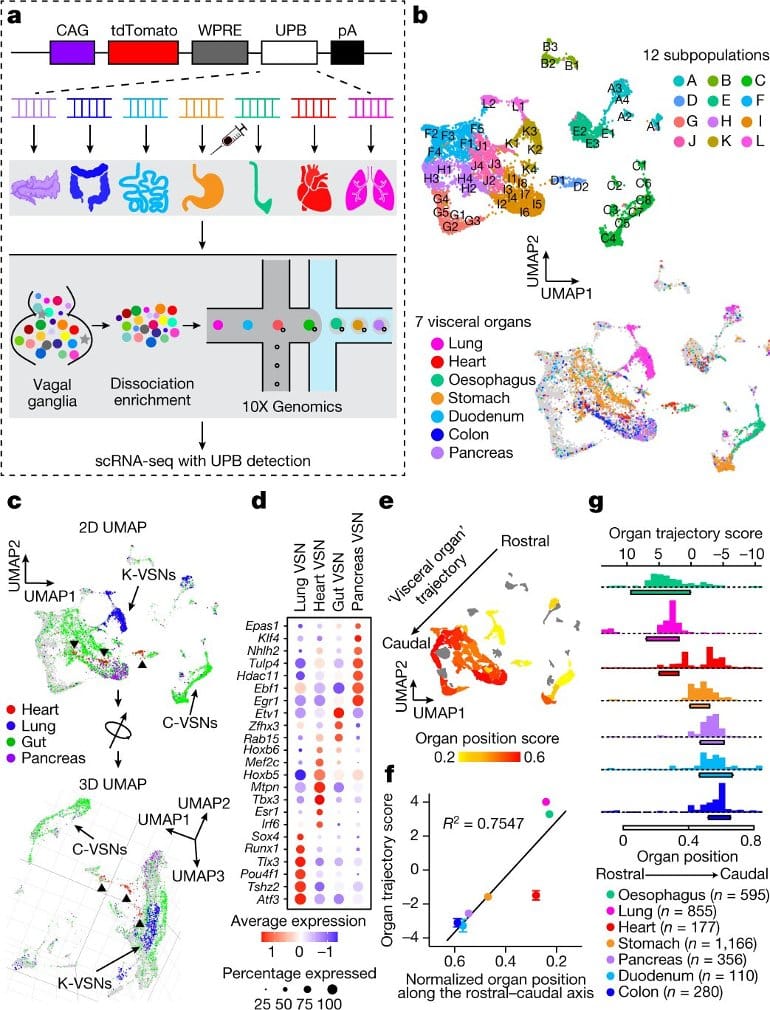
First, the researchers wanted to understand how organ information is coded within the vagus nerve. To learn more about how vagal sensory neurons are able to discriminate signals among organs, the team genetically engineered viruses to have unique barcodes composed of different foreign DNA sequences and injected them into the major visceral (internal) organs in mice.
As a result, the vagal sensory neurons that project to each organ were labeled with the distinct barcode for that organ. They then used single cell RNA sequencing technology to learn more about the genetic properties of these neurons that project to each of the seven organs.
Through this novel technology, the team discovered a “genetic trajectory,” in which neurons on one side projected to organs in the upper body like the lungs and esophagus, while neurons on the other side projected to organs in the lower abdomen.
“By looking at the genetic signature of the vagus nerve, we were able to know which organ each neuron projected to along the body’s rostro-caudal axis,” says Chang. “So in summary, our first finding is that there are genetic codes for visceral organ information in the vagus nerve.”
Researchers find a surprise
Furthermore, each of our organs is made up of individual components that have different functions. The stomach, for example, consists of tissue layers including the surface connective tissue layer, the muscular layer, and the innermost mucosa layer.
The researchers also discovered distinct genetic coding guiding the vagal sensory neurons to the different tissue layers. This coding is entirely independent of the genetic coding for organs.
“Our second finding is really surprising. No one in previous studies had even considered this,” says Chang. “By knowing these two codes, you know precisely where a particular neuron in the vagus nerve projects in the body.”
Even at the same location within the body, many kinds of changes can occur, such as mechanical changes, release of hormones, or inflammation. To better understand how the body detects these changes, the researchers developed a new technique called vagal calcium imaging transformed fluorescence in situ hybridization, or vCatFISH.
First, using in vivo calcium imaging, they visualized neuronal activity in live mice in response to various stimuli. As the mice experienced bodily changes such as stomach stretching or nutrients moving through the intestine, the researchers studied the calcium responses of the vagal ganglion to see which neurons were activated.
Using this approach, the researchers found segregated populations of neurons with similar genetic properties, each detecting a particular type of stimuli regardless of where it occurred.
“We learned that some neurons in the vagus nerve can respond to lung stretch, others respond to stomach stretch, and others can respond to intestinal nutrient perfusion,” says Chang.
“For neurons that are designed to detect stretch, for example, it doesn’t matter where the stretch happened—it could be from the lung, stomach or small intestine. In other words, neurons with the same ‘stretch’ code respond to stretches regardless of organs or tissue layers—it’s an independent, third dimension.”
New approaches to treating disease
By knowing how the vagus nerve communicates different signals to the brain, the researchers hope to be able to design tools targeting individual signal pathways.
“If we understand how the vagus nerve can control the heart, for example, this could lead to finding new ways to treat hypertension,” says Zhang.
Furthermore, vagus nerve stimulation is an effective treatment for epilepsy and depression, but researchers don’t yet understand why. By knowing which neurons are involved with specific functions, the team hopes more effective and precise treatments will follow.
“In the short term, we hope to increase the efficacy of the already existing vagus nerve stimulation approach,” says Chang. “But our long-term goal is to use our research to design treatment for many sorts of different disorders.”
About this neuroscience research news
Author: Isabella Backman
Source: Yale
Contact: Isabella Backman – Yale
Image: The image is credited to the researchers
Original Research: Open access.
“A multidimensional coding architecture of the vagal interoceptive system” by Qiancheng Zhao et al. Nature
Abstract
A multidimensional coding architecture of the vagal interoceptive system
Interoception, the ability to timely and precisely sense changes inside the body, is critical for survival.
Vagal sensory neurons (VSNs) form an important body-to-brain connection, navigating visceral organs along the rostral–caudal axis of the body and crossing the surface–lumen axis of organs into appropriate tissue layers. The brain can discriminate numerous body signals through VSNs, but the underlying coding strategy remains poorly understood.
Here we show that VSNs code visceral organ, tissue layer and stimulus modality—three key features of an interoceptive signal—in different dimensions. Large-scale single-cell profiling of VSNs from seven major organs in mice using multiplexed projection barcodes reveals a ‘visceral organ’ dimension composed of differentially expressed gene modules that code organs along the body’s rostral–caudal axis.
We discover another ‘tissue layer’ dimension with gene modules that code the locations of VSN endings along the surface–lumen axis of organs. Using calcium-imaging-guided spatial transcriptomics, we show that VSNs are organized into functional units to sense similar stimuli across organs and tissue layers; this constitutes a third ‘stimulus modality’ dimension.
The three independent feature-coding dimensions together specify many parallel VSN pathways in a combinatorial manner and facilitate the complex projection of VSNs in the brainstem.
Our study highlights a multidimensional coding architecture of the mammalian vagal interoceptive system for effective signal communication.