Scientists speed up reconstruction of connections between nerve cells more than ten-fold.
Unraveling the connectivity maps between nerve cells in brains is a huge scientific endeavor – called connectomics. The main limitation to mapping large parts of the brain is the analysis of the data obtained with electron microscopes. Berning and colleagues from the Department of Connectomics at the Frankfurt-based Max Planck Institute for Brain Research developed a novel toolset, called SegEM, which speeds up connectome analysis by more than 10-fold.
Connectomics is a relatively new research field where researchers aim to reconstruct the neuronal connectivity in parts of the brain from measured datasets. Besides the anatomical structure of neurons, this includes a reconstruction of the actual connections between the nerve cells via their synapses. As a single neuron communicates with up to thousands of other neurons and the cells are extremely densely packed, this is a difficult and extremely time-consuming endeavor. In comparison to the measurements, which already take thousands of hours, the analysis and reconstruction phase takes more than three orders of magnitude longer.
In order to make this manageable in a lifetime, two main reduction strategies are followed simultaneously. This first one is to increase the manpower – and the second strategy is to develop new algorithms to reconstruct the brain tissue data in a more automated way.
Researchers at the Department of Connectomics are already working on increasing the number of participants by developing a platform where also non-qualified personnel (e.g. students) can assist in the analysis of the connections between the neurons. The research group already recruited large populations of students to help determine the connectome of a part of the mouse retina and is currently developing the game Brainflight to have as many as people as possible participating in a similar project for the cerebral cortex.
The most recent publication in Neuron however deals with the other strategy to reduce the analysis time. Due to the complex structure and the large number of connections in a piece of cerebral cortex, a manual analysis would take up to 500.000 work hours and an investment of millions of Euros. Manuel Berning, Kevin Boergens and Moritz Helmstaedter have now discovered a way to also automatically speed up the image classification step. Berning: “By using machine learning algorithms, we were able to develop a way to automatically classify brain tissue containing all the synapses. By using these SegEM tools, we reduce the time for analysis by at least a factor of ten, bringing the analysis step closer to data generation.”
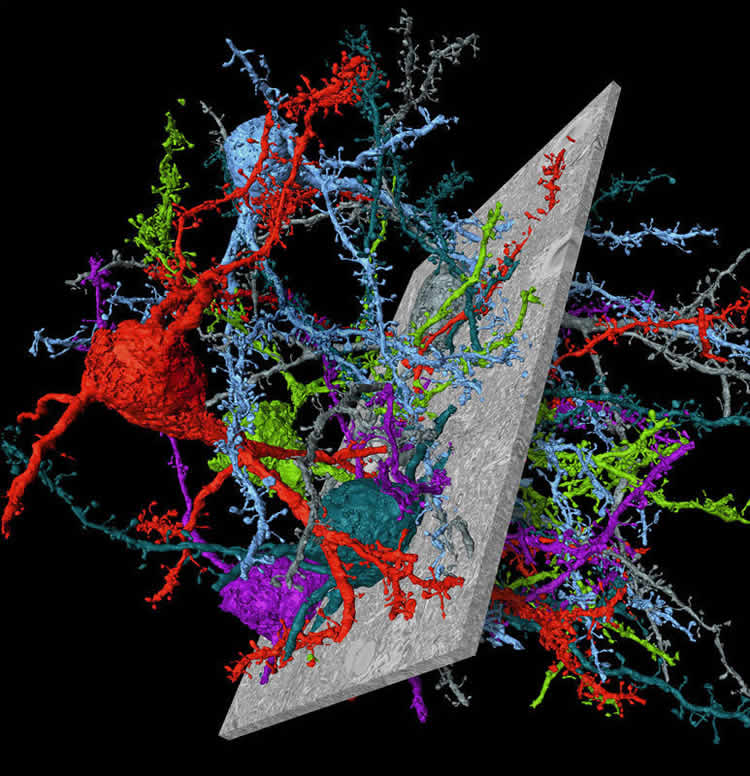
The video shows a flight along an axon in mouse cerebral cortex, showing the “skeleton” of the nerve cell followed by a display of SegEM-reconstructed nerve cells.
The researchers first trained their system with existing data sets from retina and cortex before performing an automated test of new data. Helmstaedter: “We were amazed that the new algorithm actually works extremely well for retinal and cortical data. This is real breakthrough, and an important step towards making connectome analysis a ready-to-use technique in neuroscience labs around the world.”
Source: Max Planck Institute
Image Credit: The image is credited to MPI f. Brain Research/ Berning, Boergens, Helmstaedter.
Video Source: The video is available at MaxPlanckSociety YouTube page
Original Research: Abstract for “SegEM: Efficient Image Analysis for High-Resolution Connectomicss” by Manuel Berning, Kevin M. Boergens, and Moritz Helmstaedter in Neuron. Published online September 23 2015 doi:10.1016/j.neuron.2015.09.003
Abstract
SegEM: Efficient Image Analysis for High-Resolution Connectomics
Highlights
•SegEM: semi-automated reconstruction workflow for connectomics image data
•Reconstruction speed about 10-times higher than that of alternative methods
•Applied to 3D electron microscopy data from mouse retina and cortex
•Ready-to-use analysis code for novel datasets
Summary
Progress in electron microscopy-based high-resolution connectomics is limited by data analysis throughput. Here, we present SegEM, a toolset for efficient semi-automated analysis of large-scale fully stained 3D-EM datasets for the reconstruction of neuronal circuits. By combining skeleton reconstructions of neurons with automated volume segmentations, SegEM allows the reconstruction of neuronal circuits at a work hour consumption rate of about 100-fold less than manual analysis and about 10-fold less than existing segmentation tools. SegEM provides a robust classifier selection procedure for finding the best automated image classifier for different types of nerve tissue. We applied these methods to a volume of 44 × 60 × 141 μm3 SBEM data from mouse retina and a volume of 93 × 60 × 93 μm3 from mouse cortex, and performed exemplary synaptic circuit reconstruction. SegEM resolves the tradeoff between synapse detection and semi-automated reconstruction performance in high-resolution connectomics and makes efficient circuit reconstruction in fully-stained EM datasets a ready-to-use technique for neuroscience.
“SegEM: Efficient Image Analysis for High-Resolution Connectomicss” by Manuel Berning, Kevin M. Boergens, and Moritz Helmstaedter in Neuron. Published online September 23 2015 doi:10.1016/j.neuron.2015.09.003