Looking at a brain tumor’s epigenetic signature may help guide therapy.
A comprehensive analysis of the molecular characteristics of gliomas–the most common malignant brain tumor–explains why some patients diagnosed with slow-growing (low-grade) tumors quickly succumb to the disease while others with more aggressive (high-grade) tumors survive for many years. The multinational study suggests a new way of classifying gliomas that may have a significant impact on patient management and may lead to the development of more targeted therapies.
The paper, co-led by researchers from Columbia University Medical Center (CUMC), USA, Ribeirão Preto Medical School (FMRP) at the University of São Paulo (USP), Brazil, and The University of Texas MD Anderson Cancer Center, Houston, Texas, USA, was published today in the journal Cell.
Currently, pathologists determine if a glioma is low-grade or high-grade based on the tumor tissue’s appearance under the microscope.
“While this approach is generally good at distinguishing between gliomas that are clearly very aggressive and those that are relatively slow-growing, it misses the mark in a significant percentage of cases, leading to inappropriate treatment,” said co-senior author Antonio Iavarone, MD, professor of neurology and pathology and cell biology (in the Institute for Cancer Genetics) at CUMC and a member of the Herbert Irving Comprehensive Cancer Center (HICCC) at NewYork-Presbyterian /Columbia University Medical Center. “Instead, by looking at the molecular makeup of these tumors, we now have a much more precise way of predicting which tumors are more likely to grow rapidly and can prescribe treatments accordingly.”
Other researchers have attempted to classify gliomas according to their genetic characteristics. One study found that tumors with mutations in a gene called IDH were significantly less aggressive than those without the mutation, known as IDHwildtype tumors. However, these findings did not fully explain why some patients with IDHmutant tumors fare worse than expected and some with IDHwildtype tumors fare better than expected. Other studies suggested that a glioma’s level of DNA methylation, an epigenetic process that cells use to control gene expression, might explain a tumor’s aggressiveness, but the evidence was inconclusive.
In this study, Dr. Iavarone and his colleagues analyzed 1,122 high and low grade glioma samples from the Cancer Genome Atlas, looking for epigenetic changes in the tumors’ DNA. The researchers found that the best predictor of progression in an IDHmutant glioma–the less-aggressive variety–is its level of DNA methylation. Among IDHmutant gliomas, those with a high degree of DNA methylation progressed more slowly. However, tumors with less DNA methylation, about 6 percent of the total, progressed very quickly.
“Based on their appearance under the microscope, these aggressive tumors looked very much like the other IDHmutant tumors,” said Dr. Iavarone. “But from a disease prognosis standpoint, they progressed quite similarly to the more lethal subset of IDHwildtype gliomas,” said Dr. Iavarone.
Among those with IDHwildtype gliomas–the most aggressive type–a small subset (about 6 percent) had relatively favorable clinical outcomes. The molecular characteristics of this group were similar to those of pilocytic astrocytomas, a childhood brain tumor with a relatively favorable survival rate.
“The present study advances the understanding of the glioma division by correlating each subtype of DNA methylation with a distinct clinical outcome,” said co-senior author Houtan Noushmehr, PhD, professor of epigenomics and bioinformatics at University of São Paulo and director of the OMICs and Bioinformatics lab at FMRP at Ribeirão Preto, São Paulo. “We discovered low grade and high grade gliomas mixed together within these different epigenetic subtypes. This was an unexpected finding and allowed us to further understand the progression of gliomas within the different subtypes,” said Dr. Noushmehr.
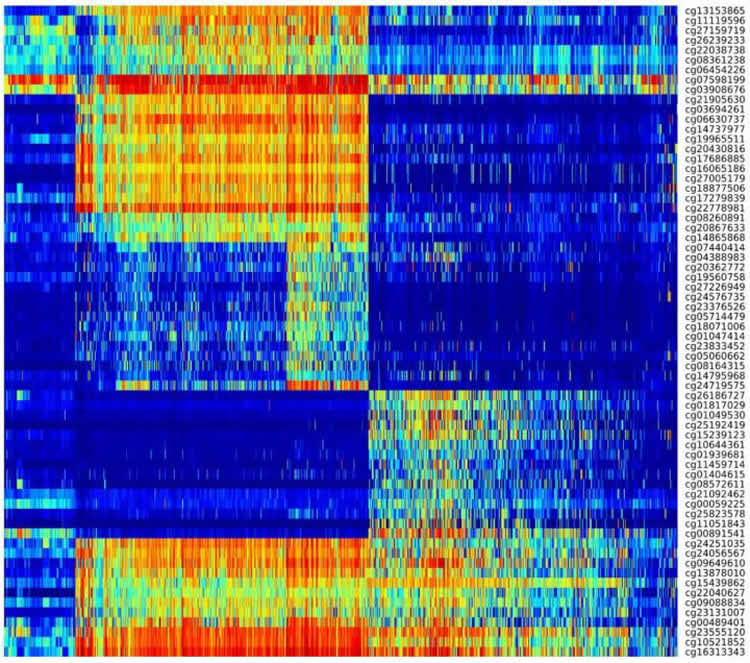
“This research has expanded our knowledge of the glioma somatic alteration landscape and emphasized the relevance of DNA methylation profiles as a method for clinical classification,” said senior co-author Roel Verhaak, PhD, associate professor of bioinformatics and computational biology MD Anderson. “These findings are an important step forward in our understanding of glioma as discrete disease subsets, and the mechanism driving glioma formation and progression.”
The paper also identified several previously unrecognized genetic alterations that may contribute to glioma development, highlighting potential new targets for drug therapy.
“This study, which focused on tumor classification, does not point to specific therapies for glioma,” said Dr. Iavarone. “But our findings will help clinicians identify subsets of patients with IDHmutant tumors who need to be treated more aggressively and those with IDHwildtype tumors who can be spared aggressive treatment.”
The study is titled, “Molecular profiling refines the classification of adult diffuse lower- and high-grade glioma.” The full list of contributors can be found in Cell.
The researchers have submitted a patent for probes that can be used to predict a glioma’s clinical outcome. The researchers declare no other conflicts of interest.
Funding: This study was supported by grants from the São Paulo Research Foundation (FAPESP) (2015/07925-5, 2015/02844-7, 2014/08321-3, 2014/02245-3), the National Institutes of Health (U24CA143883, U24CA143858, U24CA143840, U24CA143799, U24CA143835, U24CA143845, U24CA143882, U24CA143867, U24CA143866, U24CA143848, U24CA144025, U54HG003067, U54HG003079, U54HG003273, U24CA126543, U24CA126544, U24CA126546, U24CA126551, U24CA126554, U24CA126561, U24CA126563, U24CA143731, U24CA143843, P30CA016672, P50 CA127001, R01 CA190121, P01 CA085878), and the Cancer Prevention & Research Institute of Texas.
Source: Karin Eskenazi – Columbia University Medical Center
Image Credit: The image is credited to Lab of Antonio Iavarone/Columbia University Medical Center
Original Research: Abstract for “Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma” by Michele Ceccarelli, Floris P. Barthel, Tathiane M. Malta, Thais S. Sabedot, Sofie R. Salama, Bradley A. Murray, Olena Morozova, Yulia Newton, Amie Radenbaugh, Stefano M. Pagnotta, Samreen Anjum, Jiguang Wang, Ganiraju Manyam, Pietro Zoppoli, Shiyun Ling, Arjun A. Rao, Mia Grifford, Andrew D. Cherniack, Hailei Zhang, Laila Poisson, Carlos Gilberto Carlotti Jr., Daniela Pretti da Cunha Tirapelli, Arvind Rao, Tom Mikkelsen, Ching C. Lau, W.K. Alfred Yung, Raul Rabadan, Jason Huse, Daniel J. Brat, Norman L. Lehman, Jill S. Barnholtz-Sloan, Siyuan Zheng, Kenneth Hess, Ganesh Rao, Matthew Meyerson, Rameen Beroukhim, Lee Cooper, Rehan Akbani, Margaret Wrensch, David Haussler, Kenneth D. Aldape, Peter W. Laird, David H. Gutmann, show TCGA Research Network, Houtan Noushmehr, Antonio Iavarone, and Roel G.W. Verhaak in Cell. Published online January 28 2016 doi:10.1016/j.cell.2015.12.028
Abstract
Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma
Highlights
•Comprehensive molecular profiling of 1,122 adult diffuse grade II, III, and IV gliomas
•Telomere length and telomere maintenance defined by somatic alterations
•DNA methylation profiling reveals subtypes of IDH mutant and IDH-wild-type glioma
•Integrated molecular analysis of progression from low-grade to high-grade disease
Summary
Therapy development for adult diffuse glioma is hindered by incomplete knowledge of somatic glioma driving alterations and suboptimal disease classification. We defined the complete set of genes associated with 1,122 diffuse grade II-III-IV gliomas from The Cancer Genome Atlas and used molecular profiles to improve disease classification, identify molecular correlations, and provide insights into the progression from low- to high-grade disease. Whole-genome sequencing data analysis determined that ATRX but not TERT promoter mutations are associated with increased telomere length. Recent advances in glioma classification based on IDH mutation and 1p/19q co-deletion status were recapitulated through analysis of DNA methylation profiles, which identified clinically relevant molecular subsets. A subtype of IDH mutant glioma was associated with DNA demethylation and poor outcome; a group of IDH-wild-type diffuse glioma showed molecular similarity to pilocytic astrocytoma and relatively favorable survival. Understanding of cohesive disease groups may aid improved clinical outcomes.
“Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma” by Michele Ceccarelli, Floris P. Barthel, Tathiane M. Malta, Thais S. Sabedot, Sofie R. Salama, Bradley A. Murray, Olena Morozova, Yulia Newton, Amie Radenbaugh, Stefano M. Pagnotta, Samreen Anjum, Jiguang Wang, Ganiraju Manyam, Pietro Zoppoli, Shiyun Ling, Arjun A. Rao, Mia Grifford, Andrew D. Cherniack, Hailei Zhang, Laila Poisson, Carlos Gilberto Carlotti Jr., Daniela Pretti da Cunha Tirapelli, Arvind Rao, Tom Mikkelsen, Ching C. Lau, W.K. Alfred Yung, Raul Rabadan, Jason Huse, Daniel J. Brat, Norman L. Lehman, Jill S. Barnholtz-Sloan, Siyuan Zheng, Kenneth Hess, Ganesh Rao, Matthew Meyerson, Rameen Beroukhim, Lee Cooper, Rehan Akbani, Margaret Wrensch, David Haussler, Kenneth D. Aldape, Peter W. Laird, David H. Gutmann, show TCGA Research Network, Houtan Noushmehr, Antonio Iavarone, and Roel G.W. Verhaak in Cell. Published online January 28 2016 doi:10.1016/j.cell.2015.12.028