Summary: Researchers have identified a dual role for a gene associated with sleep and wakefulness regulation.
Source: Kyoto University.
A long-studied factor controlling human sleep cycles actually has two forms, one that keeps a key protein stable and another that promotes its degradation.
From the heart and lungs that propel our blood, to our rumbling stomachs at lunch time and feeling of tiredness in the evening, our bodies keep steady rhythms day in and day out.
This ‘circadian clock’ is a series of tightly controlled cycles of specific amounts of proteins that make us either sleepy or wakeful. It has recently attracted public attention with the 2017 Nobel Prize for Physiology or Medicine, but numerous aspects of the clock’s functions or how it regulates our health remain a mystery.
One of the first circadian rhythm sleep disorders to be discovered was Familial Advanced Sleep Phase Syndrome, or FASPS. Patients with this condition fall asleep early in the evening, around 7 pm, and wake up in the early mornings, around 3 am, and are thus unable to adjust to standard cycles without undergoing treatment.
“FASPS is characterized by a mutation in the gene that codes for a protein essential for the circadian clock, called Period 2, or PER2,” explains corresponding author Jean-Michel Fustin from Kyoto University’s Graduate School of Pharmaceutical Science. “The stability of the PER2 protein is a key factor in determining how fast your circadian clock ticks.”
In FASPS patients, PER2 is unstable, resulting in a sped-up clock. This is due to a mutation that changes one amino acid in the protein from a serine to a glycine.
“The stability of PER2 is regulated by the phosphorylation of several critical amino acids — a common way proteins are regulated in the body. This process is conducted by other proteins called kinases,” continues Fustin. “We knew of a kinase that could destabilize PER2 — Casein kinase 1 delta, or CK1D — but we couldn’t find one that could stabilize it by phosphorylating the serine that is mutated in FASPS patients.”
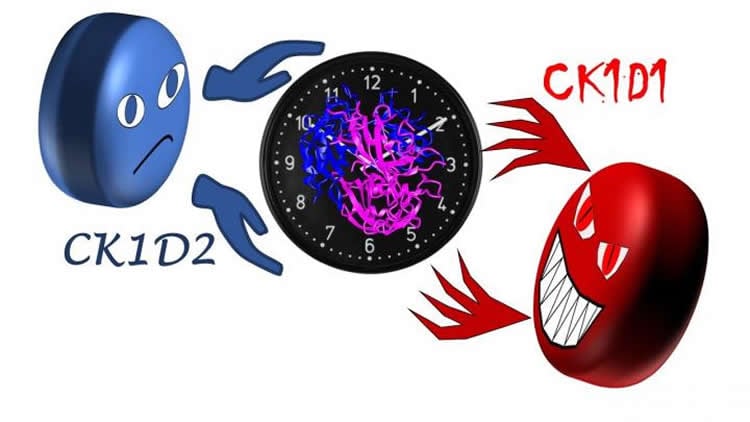
Writing in PNAS, Fustin and his collaborators explain that the stabilizing kinase was within the Ck1d gene itself all along. Their findings show that in addition to destabilizing PER2, Ck1d has another form, one that does the exact opposite.
While structurally very similar, these two kinase forms are named CK1D1 and CK1D2.
“The opposite activity of CK1D2 was completely unexpected. These two versions of this kinase can be likened to the kind Dr Jekyll — CK1D2 — and the destroyer Mr Hyde — CK1D1,” states Fustin. “While in many cases the same gene can code for different proteins, such a stark difference in function is rare.”
“Circadian clock mechanisms can be found in bacteria, insects, plants, and vertebrates. Understanding these fundamental mechanisms allows us to understand our relationship with the rhythmic environment,” concludes co-corresponding author Hitoshi Okamura. “Our discoveries indicate that the circadian clock can be adjusted between these kinases, and provides new targets for the treatment of circadian disorders.”
Funding: Japan Science and Technology Agency, Kato Memorial Bioscience Foundation, Senri Life Science Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Kobayashi International Scholarship Foundation, and others funded this study.
Source: Raymond Kunikane Terhune – Kyoto University
Publisher: Organized by NeuroscienceNews.com.
Image Source: NeuroscienceNews.com image is credited to Kyoto University / Jean-Michel Fustin.
Original Research: Open access research for “Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock” by Jean-Michel Fustin, Rika Kojima, Kakeru Itoh, Hsin-Yi Chang, Shiqi Ye, Bowen Zhuang, Asami Oji, Shingo Gibo, Rajesh Narasimamurthy, David Virshup, Gen Kurosawa, Masao Doi, Ichiro Manabe, Yasushi Ishihama, Masahito Ikawa, and Hitoshi Okamura in PNAS. Published May 15 2018.
doi:10.1073/pnas.1721371115
[cbtabs][cbtab title=”MLA”]Kyoto University”New Gears in Your Sleep Clock.” NeuroscienceNews. NeuroscienceNews, 11 July 2018.
<https://neurosciencenews.com/circadian-clock-gears-9553/>.[/cbtab][cbtab title=”APA”]Kyoto University(2018, July 11). New Gears in Your Sleep Clock. NeuroscienceNews. Retrieved July 11, 2018 from https://neurosciencenews.com/circadian-clock-gears-9553/[/cbtab][cbtab title=”Chicago”]Kyoto University”New Gears in Your Sleep Clock.” https://neurosciencenews.com/circadian-clock-gears-9553/ (accessed July 11, 2018).[/cbtab][/cbtabs]
Abstract
Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock
The N6-methylation of internal adenosines (m6A) in mRNA has been quantified and localized throughout the transcriptome. However, the physiological significance of m6A in most highly methylated mRNAs is unknown. It was demonstrated previously that the circadian clock, based on transcription-translation negative feedback loops, is sensitive to the general inhibition of m6A. Here, we show that the Casein Kinase 1 Delta mRNA (Ck1δ), coding for a critical kinase in the control of circadian rhythms, cellular growth, and survival, is negatively regulated by m6A. Inhibition of Ck1δ mRNA methylation leads to increased translation of two alternatively spliced CK1δ isoforms, CK1δ1 and CK1δ2, uncharacterized until now. The expression ratio between these isoforms is tissue-specific, CK1δ1 and CK1δ2 have different kinase activities, and they cooperate in the phosphorylation of the circadian clock protein PER2. While CK1δ1 accelerates the circadian clock by promoting the decay of PER2 proteins, CK1δ2 slows it down by stabilizing PER2 via increased phosphorylation at a key residue on PER2 protein. These observations challenge the previously established model of PER2 phosphorylation and, given the multiple functions and targets of CK1δ, the existence of two isoforms calls for a re-evaluation of past research when CK1δ1 and CK1δ2 were simply CK1δ.