Researchers at the Howard Hughes Medical Institute’s Janelia Research Campus have uncovered unexpected diversity within a single, canonical cell type of the mammalian brain. Their analysis of cells called CA1 pyramidal neurons shows that for many genes, expression levels vary with the cells’ anatomical positions, gradually building or weakening along the length of the hippocampus.
CA1 pyramidal neurons act as the output cells of a well-studied circuit in the hippocampus. The cells all share a characteristic shape and reside in the same structure in the brain. But according to the new research, published online January 14, 2016, in the journal Neuron, gene expression varies as much among CA1 pyramidal cells as it does between these cells and other hippocampal cells having entirely different forms and functions.
“This study has really emphasized the extreme variability of gene expression in what appears to be a gradual, continuous fashion along the dorsal-ventral axis of the hippocampus,” says Nelson Spruston, Scientific Program Director and Laboratory Head at Janelia who led the work. “Our findings establish a new organizational principle influencing cellular identity, in which neuronal identity can strongly change in a continuous fashion across a large and spatially distributed population of neurons.”
The hippocampus receives and processes signals from many parts of the brain, contributing to a range of neural functions including memory and spatial navigation. Its role varies along the length of its banana-shaped structure, with one end heavily involved in cognitive function and the other contributing more to emotional processing. This may be due to the kinds of signals that are received at various points along the length of the structure, but scientists have also wondered whether intrinsic properties of cells in different regions contribute to the apparent division of labor within the hippocampus.
Mark Cembrowski, a research scientist in Spruston’s lab, investigated this possibility by comparing gene activity between different CA1 pyramidal cells from mice. Using RNA-seq, a technology that lets researchers measure the expression levels of every gene in the genome, Cembrowski compared groups of cells that he had isolated from different regions of the hippocampus. Based on work from other labs, Cembrowski says, he expected he might find a dozen or so genes that were expressed differently in separate parts of the hippocampus. Instead, he found hundreds.
The differences were most dramatic along the length (or “long axis”) of the hippocampus, so Cembrowski focused his attention there. The list of genes that were differentially expressed between the hippocampus’s two ends–its dorsal and ventral regions—was long and diverse, indicating likely effects on cells’ physical properties as well as their connections with other cells. Ultimately, these properties determine how a neuron processes information. “We found hits on essentially every functional category of genes that any neuroscientist would care about,” Cembrowski says.
Cembrowski also compared the CA1 pyramidal cells to another group of cells in the hippocampus, CA3 pyramidal cells. Despite sharing a similar name, Cembrowski says, “what CA3 pyramidal cells do in the hippocampus is totally different. They differ in their morphology, their electrophysiology, their inputs and outputs. They have different types of computational strategies.” But when it came to gene expression, Cembrowski’s analysis showed, CA3 pyramidal cells are no more different from CA1 pyramidal neurons than CA1 pyramidal neurons are from one another.
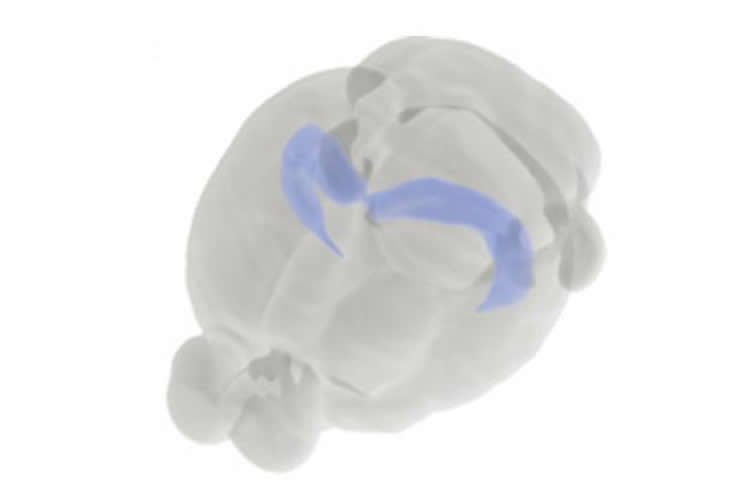
To better understand how CA1 pyramidal cells with different properties are organized within the hippocampus, the scientists then focused on a subset of the genes they knew were more active at one end of the hippocampus than the other. Using a method called in situ hybridization to label samples of brain tissue, Cembrowski could visualize exactly where in the hippocampus a particular gene was switched on. Experiments conducted in Spruston’s lab were supplemented with in situ hybridization data from a publicly available brain atlas produced by the Allen Institute for Brain Science.
That data revealed that the activity of each gene changed gradually along the axis, with each gene strengthening or weakening expression of differentially expressed genes. “What that means is that each cell has a fairly unique complement of genes that is determined by its position along this dorsal-ventral axis,” Spruston says.
“As neuroscientists, we like to think about cells being grouped into discrete, non-overlapping bits,” says Cembrowski. “But we didn’t find any evidence of discrete, well defined cell types. Rather, at least when it comes to geography, these graded changes are the organizational principle [for CA1 pyramidal cells].” Thus, to classify CA1 pyramidal cells in ways that that will help neuroscientists better understand their function, Spruston says, researchers may need to focus on cellular features, such as connectivity with other parts of the brain.
Source: Howard Hughes Medical Institute Janelia Research Campus
Image Credit: The image is credited to Brain Explorer 2, Allen Brain Atlas
Original Research: Abstract for “Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons” by Mark S. Cembrowski, Julia L. Bachman, Lihua Wang, Ken Sugino, Brenda C. Shields, and Nelson Spruston in Neuron. Published online January 14 2016 doi:10.1016/j.neuron.2015.12.013
Abstract
Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons
Highlights
•CA1 pyramidal cells (CA1 PCs) exhibit pronounced transcriptional heterogeneity
•Greatest heterogeneity is across long (dorsal-ventral; D-V) axis
•D-V differences in CA1 PCs are graded and continuous
•D-V differences within CA1 are comparable to differences across cell classes
Summary
Tissue and organ function has been conventionally understood in terms of the interactions among discrete and homogeneous cell types. This approach has proven difficult in neuroscience due to the marked diversity across different neuron classes, but it may be further hampered by prominent within-class variability. Here, we considered a well-defined canonical neuronal population—hippocampal CA1 pyramidal cells (CA1 PCs)—and systematically examined the extent and spatial rules of transcriptional heterogeneity. Using next-generation RNA sequencing, we identified striking variability in CA1 PCs, such that the differences within CA1 along the dorsal-ventral axis rivaled differences across distinct pyramidal neuron classes. This variability emerged from a spectrum of continuous gene-expression gradients, producing a transcriptional profile consistent with a multifarious continuum of cells. This work reveals an unexpected amount of variability within a canonical and narrowly defined neuronal population and suggests that continuous, within-class heterogeneity may be an important feature of neural circuits.
“Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons” by Mark S. Cembrowski, Julia L. Bachman, Lihua Wang, Ken Sugino, Brenda C. Shields, and Nelson Spruston in Neuron. Published online January 14 2016 doi:10.1016/j.neuron.2015.12.013