Summary: A new study describes the genomic landscape of schwannoma brain cancers.
Source: UHN.
Researchers from the University Health Network (UHN); Toronto Western Neurosurgery Division and MacFeeters Hamilton Neuro-oncology Program at the Princess Margaret Cancer Centre have described the genomic landscape of schwannomas in a paper published online today in Nature Genetics.
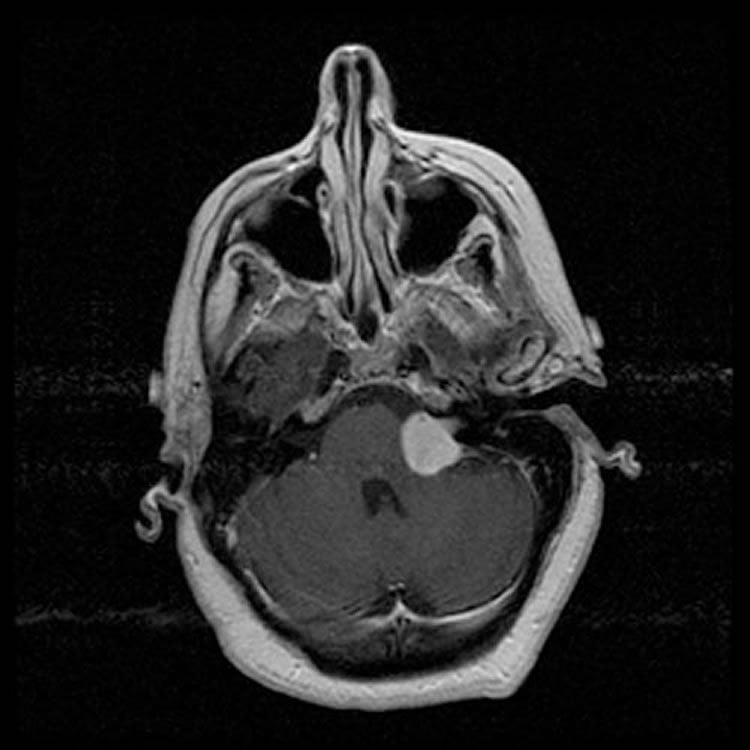
Schwannomas are one of the most common posterior fossa brain tumours and the most common spinal tumour. They stem from schwann cells, which are the cells that surround nerves and are critical in nerve function. Schwannomas commonly affect one of the cranial nerves – typically the acoustic/vestibular nerve (associated with hearing), as well as nerves leaving the spinal cord towards various sites in the body. While schwannomas are benign tumors, often they can cause debilitating symptoms depending on their location and size. Schwannomas can cause significant long-lasting neurological deficits due to a compromising of the nerve they arise from or a compression of the brain and spinal cord structures they compress. In rare cases, there can be swelling of the brain or death depending on the location of the tumours. Schwannomas can occur either sporadically or in the context of a genetic predisposition called Neurofibromatosis.
The team performed a comprehensive and integrative multi-platform molecular analysis of brain and spinal schwannomas, describing new mutations in genes that are associated with the development of schwannomas. Principal Investigator Dr. Gelareh Zadeh says that this work has identified new molecular alterations as drivers of schwannoma growth that have not previously been known and notes that the most exciting aspect of the study is that the results can change the course of treatment for some schwannomas. Dr. Zadeh, a Neurosurgeon at Toronto Western Hospital at UHN, holds the Wilkins Family Chair in Brain Tumour Research, is a Scientist, at the MacFeeters–Hamilton Neuro-oncology Program at Princess Margaret Cancer Centre and is the Head of Surgical Oncology at UHN. She is an Associate Professor in the Department of Surgery at the University of Toronto.
“Our work underlines the value of translational science carried out best through an international team approach,” says Dr. Zadeh. “We analyzed clinical samples using state-of-the-art molecular techniques in collaboration with expertise within UHN and internationally to establish fusion breakpoints, mutations, and targeted molecular sequencing of tumour samples. The combined effort from these investigators has allowed them to create the possibility that patients with schwannomas can benefit from a personalized approach to their tumour when treatment options are being considered. I must recognize Sameer Agnihotri, who is the first author on this elegant paper describing all of the work necessary to achieve this result, and Dr. Kenneth Aldape, who has been instrumental in guiding the focus of the project.”
The team profiled 125 clinically annotated schwannoma tumour samples and, using molecular testing, was able to identify subsets of tumours. One subset in particular showed a novel gene fusion (abnormal joining of 2 genes together). The two genes, called HTRA1 and SH3PXD2A, are physically separated in normal cells, but in a subset of schwannoma tumors, show a rearrangement, whereby a portion of one gene is joined to a portion of the other, creating a fusion gene. The authors demonstrate that the fusion gene has cellular functions consistent with the promotion of tumour development, suggesting that it functions as a “driver” of tumorigenesis. Since the fusion is unique to tumor cells, it represents an ideal candidate for therapeutic targeting, since abolishing its function would represent a tumour-specific approach that would spare normal cells in the body. The fusion occurs in the same site future work by the investigators will focus on targeting this abnormal fusion gene for diagnostic and therapeutic development.
“Personalized medicine describes an approach that requires the understanding of a patient’s tumour cell structure,” says Dr. Aldape, the paper’s co-Senior Author. “This work demonstrates that a molecular understanding of tumours such as schwannomas will redefine the existing understanding and approaches toward the diagnosis and classification of these tumours. With this knowledge we can move on to develop individual treatment approaches to the various genetic mutations, in particular the fusion proteins.” Dr Aldape is a molecular neuro-pathologist, Senior Scientist and Director of MacFeeters-Hamilton Neuro-oncology Program at Princess Margaret Cancer Centre.
Dr. Sameer Agnihotri, first author of the study, highlights the importance of the fusion protein through a series of elegant studies that demonstrate how the fusion promotes the development of schwannomas via its effect on cell proliferation and cell invasion. He notes that “our work demonstrates that tumours are dependent on this fusion protein, making it an ideal therapeutic target for a subset of patients with treatment-refractory schwannoma.” Dr. Agnihotri is a Scientific Associate in the MacFeeters-Hamilton Neuro-oncology Program.
Funding: Drs. Zadeh, Aldape and Agnihotri have a research program funded by Canadian Institute for Health Research (CHIR), Princess Margaret Cancer Foundation, the Toronto General and Toronto Western Hospital Foundation through the Wilkins Family Chair in Brain Tumour Research, the Terry Fox Research Institute, Elizabeth Raab Neurofibromatosis Program and the MacFeeters-Hamilton Neuro-Oncology Research Program.
Source: Jarrett Churchill – UHN
Image Source: NeuroscienceNews.com image is credited to RadsWiki and is licensed CC By SA 3.0.
Video Source: The video is credited to UHNToronto.
Original Research: Abstract for “The genomic landscape of schwannoma” by Sameer Agnihotri, Shahrzad Jalali, Mark R Wilson, Arnavaz Danesh, Mira Li, George Klironomos, Jonathan R Krieger, Alireza Mansouri, Osaama Khan, Yasin Mamatjan, Natalie Landon-Brace, Takyee Tung, Mark Dowar, Tiantian Li, Jeffrey P Bruce, Kelly E Burrell, Peter D Tonge, Amir Alamsahebpour, Boris Krischek, Pankaj Kumar Agarwalla, Wenya Linda Bi, Ian F Dunn, Rameen Beroukhim, Michael G Fehlings, Vera Bril, Stefano M Pagnotta, Antonio Iavarone, Trevor J Pugh, Kenneth D Aldape and Gelareh Zadeh in Nature Genetics. Published online October 10 2016 doi:10.1038/ng.3688
[cbtabs][cbtab title=”MLA”]UHN. “Researchers Map Genomic Landscape of Schwannoma Brain Cancer.” NeuroscienceNews. NeuroscienceNews, 10 October 2016.
<https://neurosciencenews.com/schwannoma-cancer-genetics-5258/>.[/cbtab][cbtab title=”APA”]UHN. (2016, October 10). Researchers Map Genomic Landscape of Schwannoma Brain Cancer. NeuroscienceNews. Retrieved October 10, 2016 from https://neurosciencenews.com/schwannoma-cancer-genetics-5258/[/cbtab][cbtab title=”Chicago”]UHN. “Researchers Map Genomic Landscape of Schwannoma Brain Cancer.” https://neurosciencenews.com/schwannoma-cancer-genetics-5258/ (accessed October 10, 2016).[/cbtab][/cbtabs]
Abstract
The genomic landscape of schwannoma
Schwannomas are common peripheral nerve sheath tumors that can cause debilitating morbidities. We performed an integrative analysis to determine genomic aberrations common to sporadic schwannomas. Exome sequence analysis with validation by targeted DNA sequencing of 125 samples uncovered, in addition to expected NF2 disruption, recurrent mutations in ARID1A, ARID1B and DDR1. RNA sequencing identified a recurrent in-frame SH3PXD2A-HTRA1 fusion in 12/125 (10%) cases, and genomic analysis demonstrated the mechanism as resulting from a balanced 19-Mb chromosomal inversion on chromosome 10q. The fusion was associated with male gender predominance, occurring in one out of every six men with schwannoma. Methylation profiling identified distinct molecular subgroups of schwannomas that were associated with anatomical location. Expression of the SH3PXD2A-HTRA1 fusion resulted in elevated phosphorylated ERK, increased proliferation, increased invasion and in vivo tumorigenesis. Targeting of the MEK-ERK pathway was effective in fusion-positive Schwann cells, suggesting a possible therapeutic approach for this subset of tumors.
“The genomic landscape of schwannoma” by Sameer Agnihotri, Shahrzad Jalali, Mark R Wilson, Arnavaz Danesh, Mira Li, George Klironomos, Jonathan R Krieger, Alireza Mansouri, Osaama Khan, Yasin Mamatjan, Natalie Landon-Brace, Takyee Tung, Mark Dowar, Tiantian Li, Jeffrey P Bruce, Kelly E Burrell, Peter D Tonge, Amir Alamsahebpour, Boris Krischek, Pankaj Kumar Agarwalla, Wenya Linda Bi, Ian F Dunn, Rameen Beroukhim, Michael G Fehlings, Vera Bril, Stefano M Pagnotta, Antonio Iavarone, Trevor J Pugh, Kenneth D Aldape and Gelareh Zadeh in Nature Genetics. Published online October 10 2016 doi:10.1038/ng.3688