Summary: Using data from neuroimaging, a machine learning algorithm has identified key functional neural connections that could serve as a biomarker in the diagnosis of major depressive disorder.
Source: PLOS
Using machine learning, researchers have identified novel, distinct patterns of coordinated activity between different parts of the brain in people with major depressive disorder–even when different protocols are used to detect these brain networks. Ayumu Yamashita of Advanced Telecommunications Research Institutes International in Kyoto, Japan, and colleagues present these findings in the open-access journal PLOS Biology.
While major depression is usually straightforward to diagnose, a better understanding of the brain networks associated with depression could improve treatment strategies. Machine-learning algorithms can be applied to data on brain activity in people with depression in order to find such associations. However, most studies have focused only on specific subtypes of depression, or they have not accounted for the differences in brain imaging protocols between healthcare institutions.
To address these challenges, Yamashita and colleagues used machine learning to analyze brain network data from 713 people, 149 of whom had major depression. These data had been collected using a technique called resting-state functional MRI (rs-fMRI), which detects brain activity and produces images that reveal coordinated activity, or “functional connections,” between different parts of the brain. The imaging had been performed at different institutions using different protocols.
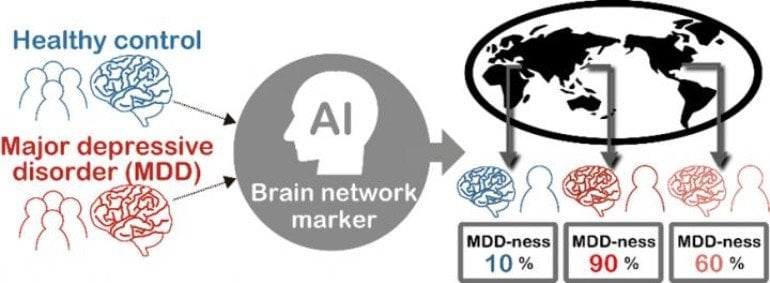
The machine-learning method identified key functional connections in the imaging data that could serve as a brain network signature for major depression. Indeed, when the researchers applied that new signature to rs-fMRI data collected at different institutions from 521 other people, they achieved 70 percent accuracy in identifying which of those new people had major depressive disorder.
The researchers hope that their new brain network signature, which can be applied across different imaging protocols, could serve as a foundation for discovering brain network patterns associated with subtypes of depression, and to reveal relationships between depression and other disorders. A better understanding of brain network connections in major depression could help match patients to effective treatments and inform development of new treatments.
About this machine learning research news
Source: PLOS
Contact: Ayumu Yamashita – PLOS
Image: The image is credited to Ayumu Yamashita
Original Research: Open access.
“Generalizable brain network markers of major depressive disorder across multiple imaging sites” by Ayumu Yamashita, Yuki Sakai, Takashi Yamada, Noriaki Yahata, Akira Kunimatsu, Naohiro Okada, Takashi Itahashi, Ryuichiro Hashimoto, Hiroto Mizuta, Naho Ichikawa, Masahiro Takamura, Go Okada, Hirotaka Yamagata, Kenichiro Harada, Koji Matsuo, Saori C. Tanaka, Mitsuo Kawato, Kiyoto Kasai, Nobumasa Kato, Hidehiko Takahashi, Yasumasa Okamoto, Okito Yamashita, Hiroshi Imamizu. PLOS Biology
Abstract
Generalizable brain network markers of major depressive disorder across multiple imaging sites
Many studies have highlighted the difficulty inherent to the clinical application of fundamental neuroscience knowledge based on machine learning techniques. It is difficult to generalize machine learning brain markers to the data acquired from independent imaging sites, mainly due to large site differences in functional magnetic resonance imaging. We address the difficulty of finding a generalizable marker of major depressive disorder (MDD) that would distinguish patients from healthy controls based on resting-state functional connectivity patterns. For the discovery dataset with 713 participants from 4 imaging sites, we removed site differences using our recently developed harmonization method and developed a machine learning MDD classifier. The classifier achieved an approximately 70% generalization accuracy for an independent validation dataset with 521 participants from 5 different imaging sites. The successful generalization to a perfectly independent dataset acquired from multiple imaging sites is novel and ensures scientific reproducibility and clinical applicability.