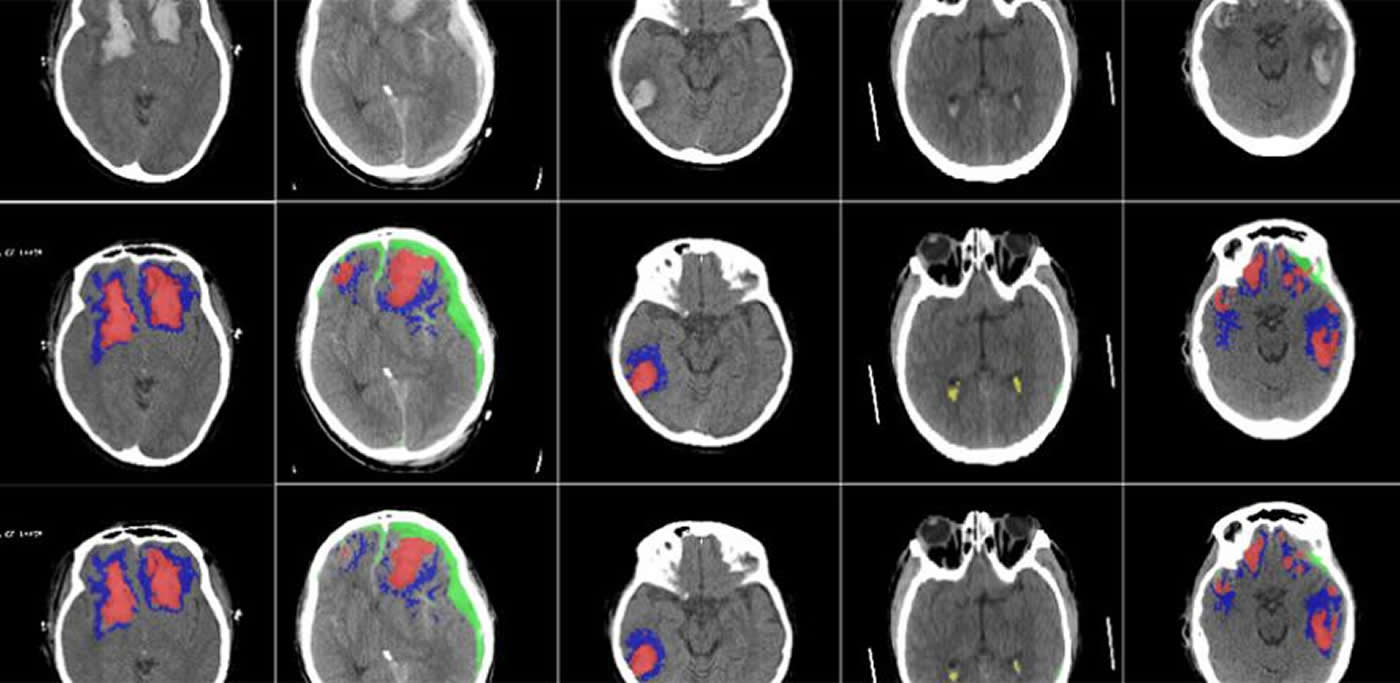
Summary: Artificial intelligence technology that uses CT brain scan images is successfully able to detect, segments, quantify, and differentiate between different types of brain lesions.
Source: University of Cambridge
Researchers have developed an AI algorithm that can detect and identify different types of brain injuries.
The researchers, from the University of Cambridge and Imperial College London, have clinically validated and tested the AI on large sets of CT scans, and found that it was successfully able to detect, segment, quantify and differentiate different types of brain lesions.
The results, reported in The Lancet Digital Health, could be useful in large-scale research studies, for developing more personalised treatments for head injuries and, with further validation, could be useful in certain clinical scenarios, such as those where radiological expertise is at a premium.
Head injury is a huge public health burden around the world and affects up to 60 million people each year. It is the leading cause of mortality in young adults. When a patient has had a head injury, they are usually sent for a CT scan to check for blood in or around the brain, and to help determine whether surgery is required.
“CT is an incredibly important diagnostic tool, but it’s rarely used quantitatively,” said co-senior author Professor David Menon, from Cambridge’s Department of Medicine. “Often, much of the rich information available in a CT scan is missed, and as researchers, we know that the type, volume and location of a lesion on the brain are important to patient outcomes.”
Different types of blood in or around the brain can lead to different patient outcomes, and radiologists will often make estimates in order to determine the best course of treatment.
“Detailed assessment of a CT scan with annotations can take hours, especially in patients with more severe injuries,” said co-first author Dr. Virginia Newcombe, also from Cambridge’s Department of Medicine. “We wanted to design and develop a tool that could automatically identify and quantify the different types of brain lesions so that we could use it in research and explore its possible use in a hospital setting.”
The researchers developed a machine learning tool based on an artificial neural network. They trained the tool on more than 600 different CT scans, showing brain lesions of different sizes and types. They then validated the tool on an existing large dataset of CT scans.
The AI was able to classify individual parts of each image and tell whether it was normal or not. This could be useful for future studies in how head injuries progress, since the AI may be more consistent than a human at detecting subtle changes over time.
“This tool will allow us to answer research questions we couldn’t answer before,” said Newcombe. “We want to use it on large datasets to understand how much imaging can tell us about the prognosis of patients.”
“We hope it will help us identify which lesions get larger and progress, and understand why they progress, so that we can develop more personalised treatment for patients in future,” said Menon.
While the researchers are currently planning to use the AI for research only, they say with proper validation, it could also be used in certain clinical scenarios, such as in resource-limited areas where there are few radiologists.
In addition, the researchers say that it could have a potential use in emergency rooms, helping get patients home sooner. Of all the patients who have a head injury, only between 10 and 15% have a lesion that can be seen on a CT scan. The AI could help identify these patients who need further treatment, so those without a brain lesion can be sent home, although any clinical use of the tool would need to be thoroughly validated.
The ability to analyse large datasets automatically will also enable the researchers to solve important clinical research questions that have previously been difficult to answer, including the determination of relevant features for prognosis which in turn may help target therapies.
About this neuroscience research article
Source:
University of Cambridge
Media Contacts:
Sarah Collins – University of Cambridge
Image Source:
The image is credited to University of Cambridge.
Original Research: Open access
“Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study”. by Virginia Newcombe et al.
The Lancet Digital Health doi:10.1016/S2589-7500(20)30085-6
Abstract
Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study
Background
CT is the most common imaging modality in traumatic brain injury (TBI). However, its conventional use requires expert clinical interpretation and does not provide detailed quantitative outputs, which may have prognostic importance. We aimed to use deep learning to reliably and efficiently quantify and detect different lesion types.
Methods
Patients were recruited between Dec 9, 2014, and Dec 17, 2017, in 60 centres across Europe. We trained and validated an initial convolutional neural network (CNN) on expert manual segmentations (dataset 1). This CNN was used to automatically segment a new dataset of scans, which we then corrected manually (dataset 2). From this dataset, we used a subset of scans to train a final CNN for multiclass, voxel-wise segmentation of lesion types. The performance of this CNN was evaluated on a test subset. Performance was measured for lesion volume quantification, lesion progression, and lesion detection and lesion volume classification. For lesion detection, external validation was done on an independent set of 500 patients from India.
Findings
98 scans from one centre were included in dataset 1. Dataset 2 comprised 839 scans from 38 centres: 184 scans were used in the training subset and 655 in the test subset. Compared with manual reference, CNN-derived lesion volumes showed a mean difference of 0·86 mL (95% CI –5·23 to 6·94) for intraparenchymal haemorrhage, 1·83 mL (–12·01 to 15·66) for extra-axial haemorrhage, 2·09 mL (–9·38 to 13·56) for perilesional oedema, and 0·07 mL (–1·00 to 1·13) for intraventricular haemorrhage.
Interpretation
We show the ability of a CNN to separately segment, quantify, and detect multiclass haemorrhagic lesions and perilesional oedema. These volumetric lesion estimates allow clinically relevant quantification of lesion burden and progression, with potential applications for personalised treatment strategies and clinical research in TBI.
Funding
European Union 7th Framework Programme, Hannelore Kohl Stiftung, OneMind, NeuroTrauma Sciences, Integra Neurosciences, European Research Council Horizon 2020.
Feel Free To Share This Artificial Intelligence News.