Summary: Analyzing the gene activity map or transcriptome of different brain diseases can help identify underlying mechanisms and comorbidity. The method could also uncover new relationships among diseases and improve treatment options.
Source: PLOS
Research led by Yashar Zeighami at McGill University, Canada provides a new way to characterize brain diseases.
The study, published in PLOS Biology on April 20th shows that comparing the transcriptomes (the map of activity for all genes in the genome) related to different brain diseases can help us understand the mechanisms underlying the diseases, and why certain ones are comorbid. The method can also find new relationships among diseases, which could have an impact on clinical treatment options.
Classifying brain diseases is difficult because many have multiple genetic and environmental risk factors. On top of that, the symptoms of many brain diseases overlap. For example, Parkinson’s disease and dementia with Lewy bodies are both neurodegenerative disorders presenting with muscle tremors and rigidity, and which share some similar cognitive and behavioral symptoms.
In this and other similar cases, misdiagnosis is not uncommon and can have serious consequences for patient care. An alternative approach is to classify brain diseases based on gene activity.
The new study examined disease transcriptomes—the set of RNA transcripts from affected brain regions—for 40 different brain diseases.
The researchers found that this system could classify brain diseases into five primary groups based on where disease-risk genes were active in the brain and in which cell types. In addition to confirming known relationships among diseases, disease transcriptome analysis was able to find previously unknown relationships among diseases.
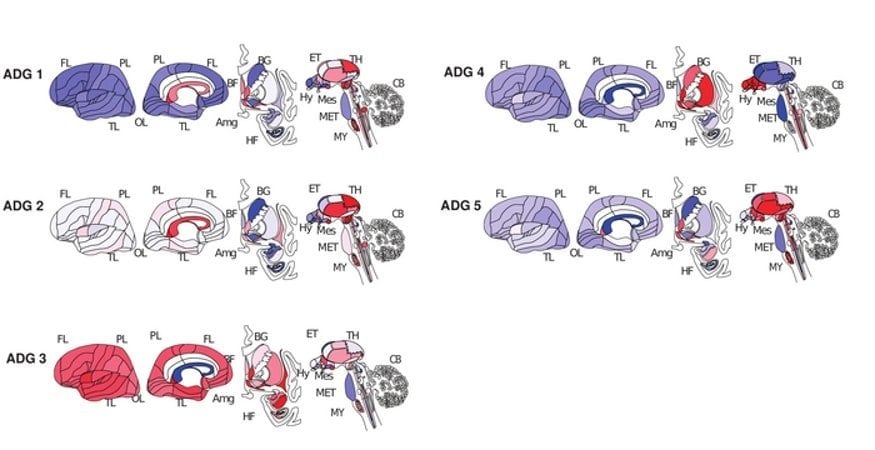
For example, language development disorders, obsessive-compulsive disorder, and temporal lobe epilepsy were all classified into group 3, meaning that despite their very different symptoms, their corresponding genes are active in the same brain regions and in the same cell types.
Brain diseases classified as neurodegenerative, movement-related, and psychiatric are the most difficult to diagnose because of the overlap in symptoms that change over time. The transcriptome is thus an additional tool that could be used for more accurate early diagnoses.
Zeighami adds, “Analysis of the transcription patterns of risk genes for human brain disease reveals characteristic expression signatures across brain anatomy. These can be used to compare and aggregate diseases, providing associations that often differ from conventional phenotypic classification.”
About this genetics and neurology research news
Author: Claire Turner
Source: PLOS
Contact: Claire Turner – PLOS
Image: The image is credited to Zeighami Y, et al., 2023, PLOS Biology, CC-BY 4.0
Original Research: Open access.
“A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases” by Yashar Zeighami et al. PLOS Biology
Abstract
A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases
Genes associated with risk for brain disease exhibit characteristic expression patterns that reflect both anatomical and cell type relationships.
Brain-wide transcriptomic patterns of disease risk genes provide a molecular-based signature, based on differential co-expression, that is often unique to that disease. Brain diseases can be compared and aggregated based on the similarity of their signatures which often associates diseases from diverse phenotypic classes.
Analysis of 40 common human brain diseases identifies 5 major transcriptional patterns, representing tumor-related, neurodegenerative, psychiatric and substance abuse, and 2 mixed groups of diseases affecting basal ganglia and hypothalamus.
Further, for diseases with enriched expression in cortex, single-nucleus data in the middle temporal gyrus (MTG) exhibits a cell type expression gradient separating neurodegenerative, psychiatric, and substance abuse diseases, with unique excitatory cell type expression differentiating psychiatric diseases.
Through mapping of homologous cell types between mouse and human, most disease risk genes are found to act in common cell types, while having species-specific expression in those types and preserving similar phenotypic classification within species.
These results describe structural and cellular transcriptomic relationships of disease risk genes in the adult brain and provide a molecular-based strategy for classifying and comparing diseases, potentially identifying novel disease relationships.






