Summary: Advances in machine learning could allow researchers to detect drug effects that may be missed through conventional statistical analysis, researchers report.
Source: UCL.
Machine learning could improve our ability to determine whether a new drug works in the brain, potentially enabling researchers to detect drug effects that would be missed entirely by conventional statistical tests, finds a new UCL study published today in Brain.
“Current statistical models are too simple. They fail to capture complex biological variations across people, discarding them as mere noise. We suspected this could partly explain why so many drug trials work in simple animals but fail in the complex brains of humans. If so, machine learning capable of modelling the human brain in its full complexity may uncover treatment effects that would otherwise be missed,” said the study’s lead author, Dr Parashkev Nachev (UCL Institute of Neurology).
To test the concept, the research team looked at large-scale data from patients with stroke, extracting the complex anatomical pattern of brain damage caused by the stroke in each patient, creating in the process the largest collection of anatomically registered images of stroke ever assembled. As an index of the impact of stroke, they used gaze direction, objectively measured from the eyes as seen on head CT scans upon hospital admission, and from MRI scans typically done 1-3 days later.
They then simulated a large-scale meta-analysis of a set of hypothetical drugs, to see if treatment effects of different magnitudes that would have been missed by conventional statistical analysis could be identified with machine learning. For example, given a drug treatment that shrinks a brain lesion by 70%, they tested for a significant effect using conventional (low-dimensional) statistical tests as well as by using high-dimensional machine learning methods.
The machine learning technique took into account the presence or absence of damage across the entire brain, treating the stroke as a complex “fingerprint”, described by a multitude of variables.
“Stroke trials tend to use relatively few, crude variables, such as the size of the lesion, ignoring whether the lesion is centred on a critical area or at the edge of it. Our algorithm learned the entire pattern of damage across the brain instead, employing thousands of variables at high anatomical resolution. By illuminating the complex relationship between anatomy and clinical outcome, it enabled us to detect therapeutic effects with far greater sensitivity than conventional techniques,” explained the study’s first author, Tianbo Xu (UCL Institute of Neurology).
The advantage of the machine learning approach was particularly strong when looking at interventions that reduce the volume of the lesion itself. With conventional low-dimensional models, the intervention would need to shrink the lesion by 78.4% of its volume for the effect to be detected in a trial more often than not, while the high-dimensional model would more than likely detect an effect when the lesion was shrunk by only 55%.
“Conventional statistical models will miss an effect even if the drug typically reduces the size of the lesion by half, or more, simply because the complexity of the brain’s functional anatomy—when left unaccounted for—introduces so much individual variability in measured clinical outcomes. Yet saving 50% of the affected brain area is meaningful even if it doesn’t have a clear impact on behaviour. There’s no such thing as redundant brain,” said Dr Nachev.
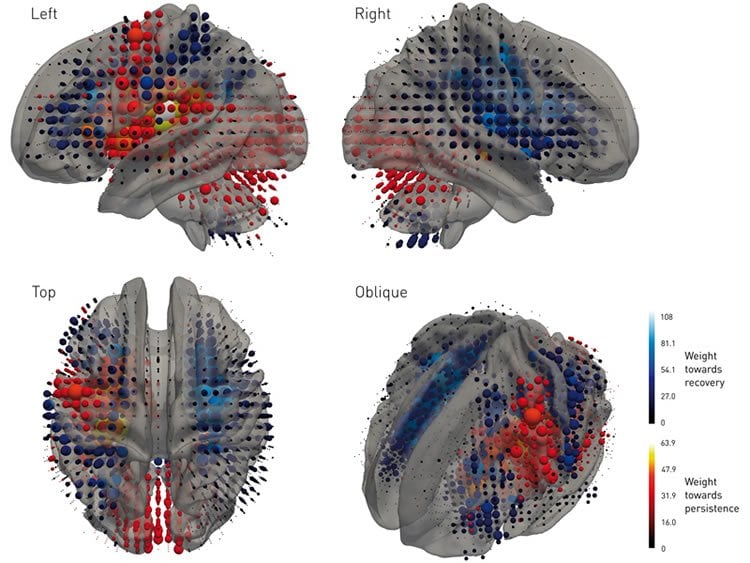
The researchers say their findings demonstrate that machine learning could be invaluable to medical science, especially when the system under study—such as the brain—is highly complex.
“The real value of machine learning lies not so much in automating things we find easy to do naturally, but formalising very complex decisions. Machine learning can combine the intuitive flexibility of a clinician with the formality of the statistics that drive evidence-based medicine. Models that pull together 1000s of variables can still be rigorous and mathematically sound. We can now capture the complex relationship between anatomy and outcome with high precision,” said Dr Nachev.
“We hope that researchers and clinicians begin using our methods the next time they need to run a clinical trial,” said co-author Professor Geraint Rees (Dean, UCL Faculty of Life Sciences).
The study was funded by Wellcome and the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Source: Chris Lane – UCL
Publisher: Organized by NeuroscienceNews.com.
Image Source: NeuroscienceNews.com image is credited to Xu et al.
Original Research: Full open access research for “High-dimensional therapeutic inference in the focally damaged human brain” by Tianbo Xu, Hans Rolf Jäger, Masud Husain, Geraint Rees, and Parashkev Nachev in Brain. Published online Noember 15 2017 doi:10.1093/brain/awx288
[cbtabs][cbtab title=”MLA”]UCL “Improving Clinical Trials With Machine Learning.” NeuroscienceNews. NeuroscienceNews, 15 November 2017.
<https://neurosciencenews.com/machine-learning-clinical-trials-7951/>.[/cbtab][cbtab title=”APA”]UCL (2017, November 15). Improving Clinical Trials With Machine Learning. NeuroscienceNews. Retrieved November 15, 2017 from https://neurosciencenews.com/machine-learning-clinical-trials-7951/[/cbtab][cbtab title=”Chicago”]UCL “Improving Clinical Trials With Machine Learning.” https://neurosciencenews.com/machine-learning-clinical-trials-7951/ (accessed November 15, 2017).[/cbtab][/cbtabs]
Abstract
High-dimensional therapeutic inference in the focally damaged human brain
Though consistency across the population renders the extraordinarily complex functional anatomy of the human brain surveyable, the inverse inference—from common functional maps to individual behaviour—is constrained by marked individual deviation from the population mean. Such inference is fundamental to the evaluation of therapeutic interventions in focal brain injury, where the impact of an induced structural change in the brain is quantified by its behavioural consequences, inevitably refracted through the lens of lesion-outcome relations. Current therapeutic evaluations do not incorporate inferences to the individual outcome derived from a detailed specification of the lesion anatomy, relying only on reductive parameters such as lesion volume and crudely discretised location. Examining 1172 patients with anatomically registered focal brain lesions, here we show that such low-dimensional models are highly insensitive to therapeutic effects. In contrast, high-dimensional models supported by machine learning dramatically improve sensitivity by leveraging complex individuating patterns in the functional architecture of the brain. The failure to replicate in humans positive interventional effects in experimental animals is thus revealed to have a remediable inferential cause, forcing a radical re-evaluation of therapeutic inference in the human brain.
“High-dimensional therapeutic inference in the focally damaged human brain” by Tianbo Xu, Hans Rolf Jäger, Masud Husain, Geraint Rees, and Parashkev Nachev in Brain. Published online Noember 15 2017 doi:10.1093/brain/awx288