Summary: Columbia team harnesses powerful technology to uncover never-before-seen details of the nervous system, demonstrating SCAPE’s broad potential to decipher the brain
Source: The Zuckerman Institute at Columbia University
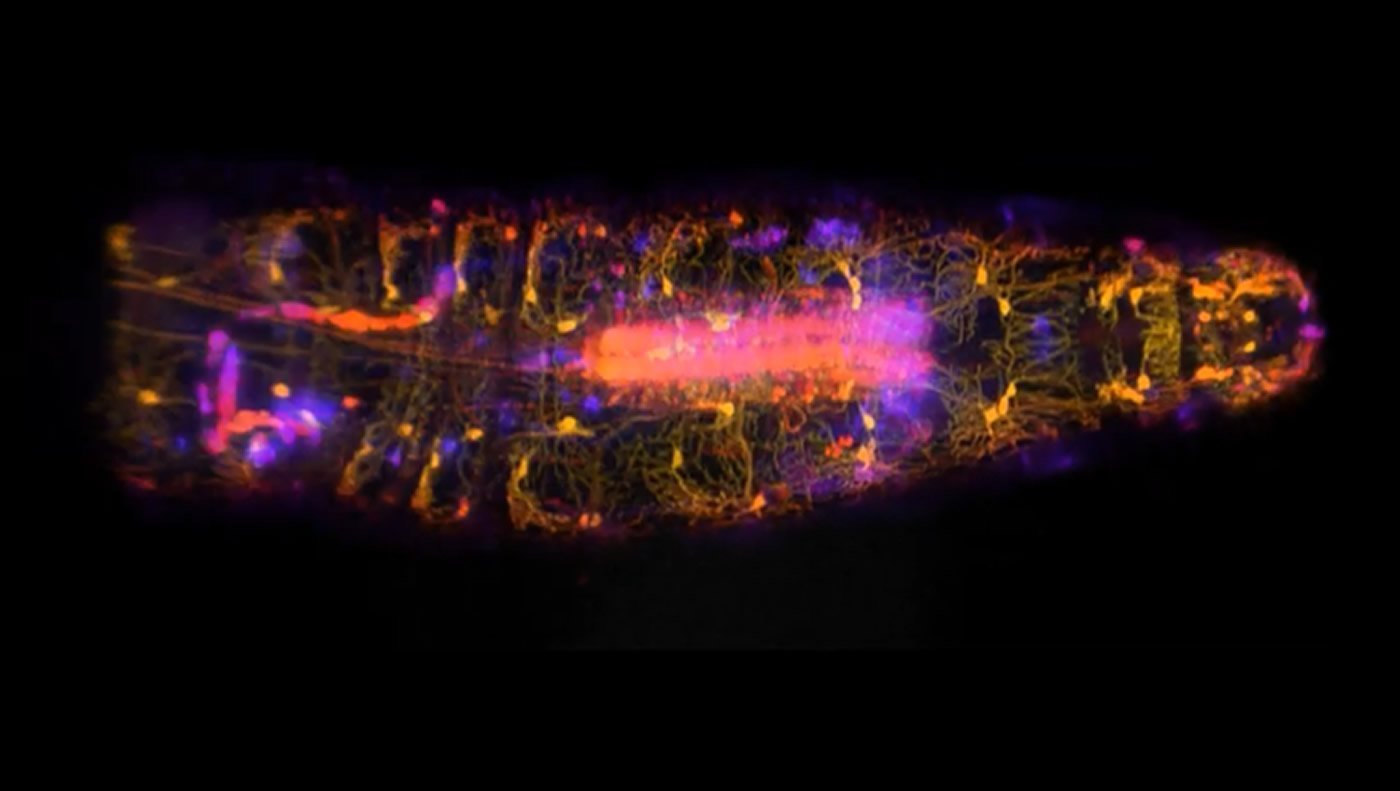
Columbia engineers and neuroscientists have joined forces to create 3D videos of individual nerve cells moving, stretching and switching on inside fruit fly larvae as they move. Data gleaned from these videos reveals how nerve cells called proprioceptive neurons work together to help the body sense where it is in space. To accomplish this feat, the researchers harnessed SCAPE, a cutting-edge microscope developed at Columbia that images neurons at lightning-fast speeds.
These findings, published in Current Biology, illustrate SCAPE’s ability to reveal the inner workings of the nervous system in unprecedented detail. By creating 3D, live action images of nerve cells in larvae as the animals crawled, SCAPE allowed the researchers to see exactly how those cells along the body wall reported movements back to the brain.
“We know that the brain receives sensory signals though electrical pulses passed along neurons, but we didn’t understand why some kinds of neurons are located in specific positions, or how particular signaling patterns represented different movements,” said Wesley Grueber, PhD, a principal investigator at Columbia’s Mortimer B. Zuckerman Mind Brain Behavior Institute and the paper’s co-senior author. “To understand this process, we needed to know what signals the neurons are sending while the larva crawled around unconstrained.”
“Although we could make larvae whose neurons were labeled with fluorescence flash as they fired, we had great difficulty imaging them,” said Rebecca Vaadia, a doctoral candidate in the Grueber lab and the paper’s co-first author. “Even our fastest microscopes required the specimen to be constrained to move unnaturally slowly, so we could never truly capture neural activity that reflected the animal’s natural, unencumbered movements until we began using SCAPE.”
“We developed SCAPE to image things in 3D really, really fast,” said Elizabeth Hillman, PhD, also a principal investigator at Columbia’s Zuckerman Institute and the paper’s co-senior author. “Working together, we quickly found that we could record nerve cells flashing inside fruit fly larvae as they crawled. Our high speeds allowed us to both measure the complex, 3D motions of the body and the neurons’ activity during those movements. And we could do so in real time.”
This imaging generated a massive amount of data. To make sense of it, the team developed algorithms that tracked each proprioceptive cell, and determined exactly when it was active as the body compressed and extended during crawling.
“We saw that the position of each cell made it sensitive to specific changes to the body’s overall shape, and when we lined up all of the neurons’ signals, we saw that they generated a detailed sequence of signals that reflected each body part’s movement,” said Wenze Li, PhD, the paper’s co-first author, who recently earned his doctorate in electrical engineering in the Hillman lab at Columbia. “It was like a beautiful machine.”
“Our experiments consistently showed that each of the proprioceptor neurons reacted differently as the larvae crawled along, an observation that could not have been made if the larvae had been constrained,” said Dr. Grueber, who is also an associate professor of physiology and cellular biophysics and of neuroscience at Columbia’s Vagelos College of Physicians and Surgeons. “We saw, in real time, how some neurons fired when the animal’s body stretched, while others fired when it compressed.”
Scientists had long hypothesized that proprioception came with redundancy because switching off one — or even some — of these neurons only made the larva crawl more slowly. However, today’s study found that each neuron has its own slightly different role to play, and is positioned precisely to sense specific aspects of the animals’ movement. The team demonstrated that this rule did not just hold for crawling, but was important for encoding more complicated movements.
“We’ve made a lot of progress in deciphering the code of these cells,” said Vaadia.
First described by Dr. Hillman and colleagues in 2015, SCAPE (swept confocally aligned planar excitation) microscopy forms 3D images of living samples by scanning them with a sheet of laser light. SCAPE’s ingenuity lies in being able to both project and detect this moving light sheet through a single, stationary objective lens. SCAPE delivers 3D-imaging speeds that are up to 500 times faster than conventional microscopes that scan objects point by point.
SCAPE is especially well-suited to imaging small organisms such as the fruit fly, since they can be quite transparent and are small enough to image their entire brains or bodies — and individual cells inside them.
“Flies, worms and fish have much simpler brains than humans, but this gives us a chance to learn how a complete nervous system actually works, cell by cell,” said Dr. Hillman, who is also a professor of biomedical engineering at Columbia Engineering and a professor of radiology at Columbia’s Vagelos College of Physicians and Surgeons. “We have good reason to believe that if we can understand these simple systems better, the lessons learned will scale up to more complex systems, including mammals.”
SCAPE’s ability to capture the brain in action comes at the perfect time as efforts such as the Fly and Human Connectome Projects are aiming to establish detailed maps of the structure and connections of the nervous system. A major goal of the BRAIN Initiative, which has funded the development of SCAPE and supported Drs. Hillman and Grueber’s collaboration, is to decipher this structure-function relationship. The ultimate goal is to leverage new tools such as SCAPE to understand exactly how the billions of connections between cells in the brain, in parallel with their complex activity, are able to generate the repertoire of complex behaviors that our brain can.
“We’ve demonstrated that SCAPE can track and map the proprioceptive neurons in a crawling fruit fly larva, and it is just the first of many studies that can now explore the fly and nervous systems of other animals,” said Dr. Li. “We can now literally label any type of cell and find out what it is doing when the animal is moving, eating or even forming a memory; the possibilities are endless.”
This paper is titled “Characterization of proprioceptive system dynamics in behaving Drosophila larvae using high-speed volumetric microscopy.” Additional contributors include Venkatakaushik Voleti and Aditi Singhania.
This research was supported by the BRAIN Initiative (5U01NS094296, UF1NS108213), the National Institutes of Health (5R01NS061908, 5T32HD00740-18), the McKnight Foundation, the Department of Defense, the Multidisciplinary University Research Initiatives Program of the Office of Naval Research (W911NF-12-1-0594), the Simons Foundation Collaboration on Global Brain and the National Science Foundation.
Elizabeth Hillman, Venkatakaushik Voleti and Wenze Li declare a financial interest in SCAPE microscopy via consulting activity and a license agreement in place between Columbia University and Leica Microsystems for commercial development of SCAPE.
Source:
The Zuckerman Institute at Columbia University
Media Contacts:
Anne Holden – Columbia University
Image Source:
High-speed SCAPE microscopy was used to capture the activity of proprioceptive neurons inside a freely moving fruit fly larvae. By creating 3D, live action images of nerve cells in larvae as the animals crawled, SCAPE allowed the researchers to see exactly how those cells along the body wall reported movements back to the brain. Credit: Wenze Li and Rebecca Vaadia/ Hillman and Grueber labs/ Columbia’s Zuckerman Institute
Original Research: Open Access
“Characterization of proprioceptive system dynamics in behaving Drosophila larvae using high-speed volumetric microscopy” by Rebecca D. Vaadia, Wenze Li, Venkatakaushik Voleti, Aditi Singhania, Elizabeth M.C. Hillman, Wesley B. Grueber in Current Biology
10.1016/j.cub.2019.01.060
Abstract
Characterization of Proprioceptive System Dynamics in Behaving Drosophila Larvae Using High-Speed Volumetric Microscopy
Highlights
• High-speed, 3D SCAPE microscopy is used to image freely moving Drosophila larvae
• Calcium activity in proprioceptive neurons is coincident with dendrite deformation
• Proprioceptor subtypes show sequential activity patterns during forward crawling
• Sensors activate in different combinations during complex turns and retractions
Summary
Proprioceptors provide feedback about body position that is essential for coordinated movement. Proprioceptive sensing of the position of rigid joints has been described in detail in several systems; however, it is not known how animals with a flexible skeleton encode their body positions. Understanding how diverse larval body positions are dynamically encoded requires knowledge of proprioceptor activity patterns in vivo during natural movement. Here we used high-speed volumetric swept confocally aligned planar excitation (SCAPE) microscopy in crawling Drosophila larvae to simultaneously track the position, deformation, and intracellular calcium activity of their multidendritic proprioceptors. Most proprioceptive neurons were found to activate during segment contraction, although one subtype was activated by extension. During cycles of segment contraction and extension, different proprioceptor types exhibited sequential activity, providing a continuum of position encoding during all phases of crawling. This sequential activity was related to the dynamics of each neuron’s terminal processes, and could endow each proprioceptor with a specific role in monitoring different aspects of body-wall deformation. We demonstrate this deformation encoding both during progression of contraction waves during locomotion as well as during less stereotyped, asymmetric exploration behavior. Our results provide powerful new insights into the body-wide neuronal dynamics of the proprioceptive system in crawling Drosophila, and demonstrate the utility of our SCAPE microscopy approach for characterization of neural encoding throughout the nervous system of a freely behaving animal.