Summary: Researchers have created a new technique that can rapidly “print’ two-dimensional arrays of cells and proteins that mimic a variety of cellular environments in the body.
Source: UC Berkeley
Like humans, cells are easily influenced by peer pressure.
Take a neural stem cell in the brain: Whether this cell remains a stem cell or differentiates into a fully formed brain cell is ultimately determined by a complex set of molecular messages the cell receives from countless neighbors. Understanding these messages is key for scientists hoping to harness these stem cells to treat neurological conditions like Alzheimer’s or Parkinson’s.
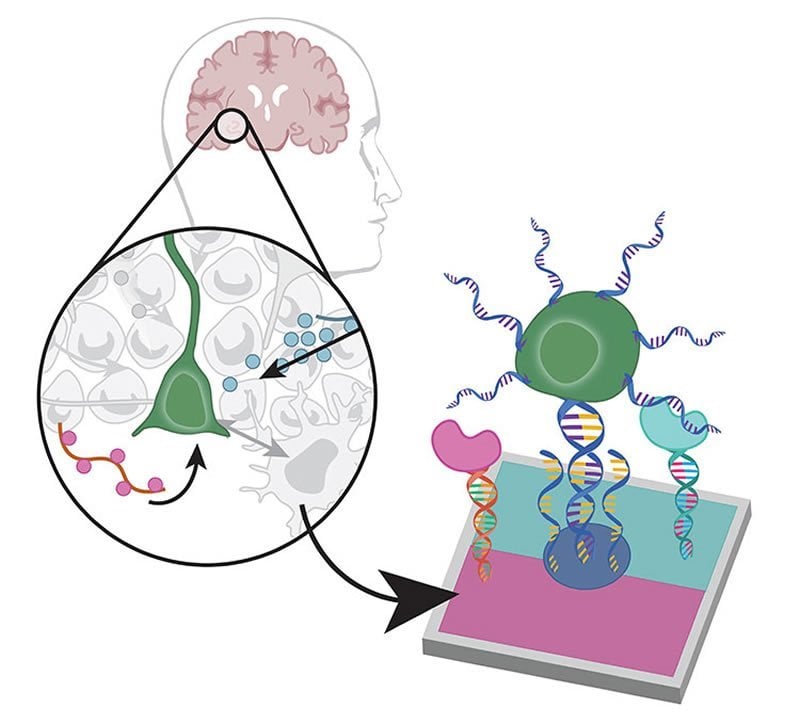
With the help of photolithography and a creative use of programmable DNA, University of California, Berkeley, researchers have created a new technique that can rapidly “print” two-dimensional arrays of cells and proteins that mimic a wide variety of cellular environments in the body — be it the brain tissue surrounding a neural stem cell, the lining of the intestine or liver or the cellular configuration inside a tumor.
This technique could help scientists develop a better understanding of the complex cell-to-cell messaging that dictates a cell’s final fate, from neural stem cell differentiating into a brain cell to a tumor cell with the potential to metastasize to an embryonic stem cell becoming an organ cell.
“What’s really powerful about this platform is you can create in vitro tissues that capture the spatial organization of cells in the body, from the intestinal lining of your digestive tract to the arrangements of different cell types in the liver,” said Olivia Scheideler, who completed the research as a graduate student at Berkeley. “I think you could apply this technique to recreate any tissue where you want to explore how cellular interactions contribute to tissue function.”
In a paper appearing today (Wednesday, March 18) in the journal Science Advances, Scheideler and her collaborators show that the new technique can be used to rapidly print intricate patterns of up to 10 different kinds cells or proteins onto a flat surface.
“Essentially, what this technique allows us to do is pattern different kinds of conditions in one shot and in a high-throughput manner,” said Lydia Sohn, Chancellor’s Professor of Mechanical Engineering at UC Berkeley and senior author of the paper. “It provides a whole range of options for what you could study, because it’s so flexible. You can pattern many different kinds of cells or proteins.”
Caught on a DNA tether
In the new technique, each cell or protein is tethered to a substrate with a short string of DNA. While similar methods have been developed that attach tethered cells or proteins one by one, the new technique takes advantage of a patterning process called photolithography to attach, or print, each type of cell protein in one quick batch, greatly speeding up the process.
“It’s like color laser printing, where you print one color and then print another,” Sohn said.
Like photography, photolithography works by exposing a coated surface or substrate to a pattern of light, which initiates a chemical reaction that dissolves the coating in the illuminated areas, leaving a templated substrate. In the new technique, the substrate is then bathed in strands of single-sided DNA, whose ends have been chemically altered to firmly latch on where the coating has been dissolved.
Each single-sided DNA strand is programmed have a specific sequence of the nucleotides adenine (A), thymine (T), guanine (G) and cytosine (C). Single-sided DNA strands with the complementary nucleotide sequence are embedded or attached to cells or proteins of interest.
Finally, the surface is washed with a mixture of cells or proteins attached to the complementary strands of single-sided DNA, which bond with the single-sided DNA already attached to the surface to form double-helix “tethers.”
“All the cells and proteins attach exactly where they should be because of the DNA programming,” Sohn said.
By repeating the process, up to 10 different kinds of cells or proteins can be tethered to the surface in an arbitrary pattern.
Conflicting messages
To demonstrate one of the many applications of the technique, Scheideler and co-author David Schaffer, Hubbard Howe Jr. Distinguished Professor of Biochemical Engineering at UC Berkeley, used the platform to study the chemical signaling that cues neural stem cells to differentiate into mature cells.
“Stem cells have programs embedded inside their DNA that tell them (to) either stay a stem cell or to differentiate into a mature cell,’” Schaffer said. “And they receive a lot of information about what to do and which programs to activate from the environment, from other cells around them. If we could learn how to make stem cells do our bidding, how to turn them into a particular cell type, then we could harness the stem cells to mass produce specialized cell types that were lost due to disease or injury.”
Neural stem cells in the brain regularly receive conflicting messages from their neighbors about how they should behave, Scheideler said. One messenger, the FGF-2 protein, tells them to make more stem cells. The other, the ephrin-B2 protein, tells them to differentiate into a mature neuron.
Scheideler used the new technique to pattern neural stem cells onto thousands of different arrays of the two proteins, FGF-2 and ephrin-B2, to see how the spatial organization of the two signals helps determine the cells’ ultimate fate.
She found that many stem cells differentiated into mature neurons, even when they were primarily in contact with FGF-2, or “stay a stem cell,” messengers. When she looked closer, however, she found that those cells that differentiated were more likely to have small, finger-like extensions, or “neurites,” that touched the ephrin-B2 or “differentiate” messengers.
“The great thing about this patterning technology is you can easily replicate these little patterns hundreds or thousands of times across a slide,” Schaffer said. “It is like running a thousand little independent experiments, each of which is a trial run to see how a stem cell listens to the cells around it. And then you can get very, very deep statistics about the various ways that it can be regulated.”
Co-authors of the paper include Chun Yang, Molly Kozminsky, Kira I. Mosher, Roberto Falcón-Banchs, Emma C. Ciminelli and Andrew W. Bremer of UC Berkeley and Sabrina A. Chern of Harvard University.
Funding: This research was supported by the National Institutes of Health through grants 1R01CA190843-01, 1R21EB019181-01A1, NIH/NCI F32 CA243354-01, and 1R21CA182375-01A1. It was also supported by the National Science Foundation Graduate Research Fellowship program, the Siebel Scholars program and a PEO Scholarship.
Source:
UC Berkeley
Media Contacts:
Kara Manke – UC Berkeley
Image Source:
The image is credited to Olivia Scheideler.
Original Research: Open access
“Recapitulating complex biological signaling environments using a multiplexed, DNA-patterning approach”. Olivia Scheideler, et al.
Science Advances doi:10.1126/sciadv.aay5696.
Abstract
Recapitulating complex biological signaling environments using a multiplexed, DNA-patterning approach
Elucidating how the spatial organization of extrinsic signals modulates cell behavior and drives biological processes remains largely unexplored because of challenges in controlling spatial patterning of multiple microenvironmental cues in vitro. Here, we describe a high-throughput method that directs simultaneous assembly of multiple cell types and solid-phase ligands across length scales within minutes. Our method involves lithographically defining hierarchical patterns of unique DNA oligonucleotides to which complementary strands, attached to cells and ligands-of-interest, hybridize. Highlighting our method’s power, we investigated how the spatial presentation of self-renewal ligand fibroblast growth factor-2 (FGF-2) and differentiation signal ephrin-B2 instruct single adult neural stem cell (NSC) fate. We found that NSCs have a strong spatial bias toward FGF-2 and identified an unexpected subpopulation exhibiting high neuronal differentiation despite spatially occupying patterned FGF-2 regions. Overall, our broadly applicable, DNA-directed approach enables mechanistic insight into how tissues encode regulatory information through the spatial presentation of heterogeneous signals.