Summary: Newly developed software allows researchers to study synaptic plasticity in dendritic spines.
Source: Max Planck Florida Institute For Neuroscience.
Researchers at Max Planck Florida Institute for Neuroscience have developed new software to study synaptic plasticity in dendritic spines.
When humans and animals learn and form memories, the physical structures of their brain cells change. Specifically, small protrusions called dendritic spines, which receive signals from other neurons, can grow and change shape indefinitely in response to stimulation. Scientists at Max Planck Florida Institute for Neuroscience (MPFI) have observed this process, known as long-term structural plasticity, in individual spines, but doing so requires substantial time and effort. A new technique, developed by MPFI researchers, automates the process to make observing and quantifying this growth far more efficient. The open-source method is available to any scientist hoping to image plasticity as it happens in dendritic spines using Scanimage. The work was published in January 2016 in the Public Library of Science journal, PLOS ONE.
Scientists working in Ryohei Yasuda’s laboratory at MPFI are working to understand how proteins facilitate the plasticity of dendritic spines, the biological basis of learning and memory. They use 2-photon microscopy, an advanced technique for live-cell imaging, and glutamate uncaging, a technique that can induce plasticity in individual spines of interest using light. This is a meticulous process, wherein a scientist must continually focus the microscope on a single dendritic spine over an extended period, often an hour or longer. Michael Smirnov, Ph.D., Post-doctoral researcher at MPFI, developed a software that allows the computer to automatically track, image, and stimulate up to five dendritic spines at a time. “We can collect the data and figure out the proteins responsible much quicker with this [program] because we can run much more robust experiments,” said Smirnov. In addition to increasing productivity, the ability to stimulate and image multiple spines in parallel greatly decreases the cost of running these experiments.
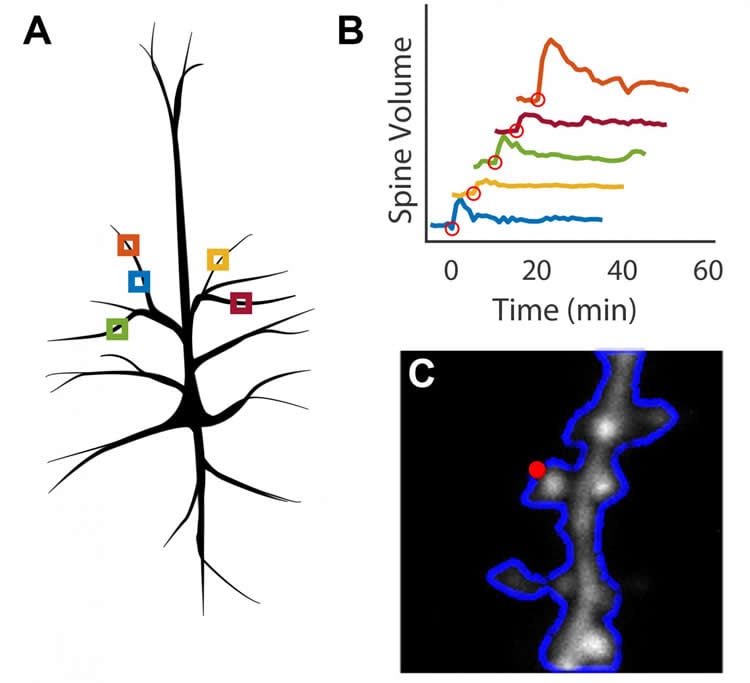
The software is a MATLAB-based module built for Scanimage, a program already commonly used in life science laboratories. It includes an electrically tunable lens in combination with a drift correction algorithm. These aspects allow the program to identify and correct for sample movement to ensure that the microscope is consistently focused on the spines of interest throughout the duration of the experiment. The interface provides an inexpensive method for automating experiments that observe up to five dendritic spines at a time, as opposed to a single spine using existing methods.
In contrast to previous open-sourced focusing programs, this one implements a highly capable and customizable focus and drift correction system to ensure that it can be used for a variety of biological applications. “The paper explains further modifications to make the process automated,” said Smirnov. “It shares the open source code, so essentially, other people from other institutes can easily pick this up and use it for themselves.”
Funding: The work was supported by the National Institutes of Health, Max Planck Florida Institute for Neuroscience, Max Planck Society.
Source: Jennifer Gutierrez – Max Planck Florida Institute For Neuroscience
Image Source: NeuroscienceNews.com image is credited to Max Planck Florida Institute for Neuroscience.
Original Research: Full open access research for “Automated Remote Focusing, Drift Correction, and Photostimulation to Evaluate Structural Plasticity in Dendritic Spines” by Michael S. Smirnov, Paul R. Evans, Tavita R. Garrett, Long Yan, and Ryohei Yasuda in PLOS ONE. Published online January 23 2017 doi:10.1371/journal.pone.0170586
[cbtabs][cbtab title=”MLA”]Max Planck Florida Institute For Neuroscience “New Software Automates Brain Imaging.” NeuroscienceNews. NeuroscienceNews, 6 February 2017.
<https://neurosciencenews.com/brain-imaging-software-6073/>.[/cbtab][cbtab title=”APA”]Max Planck Florida Institute For Neuroscience (2017, February 6). New Software Automates Brain Imaging. NeuroscienceNew. Retrieved February 6, 2017 from https://neurosciencenews.com/brain-imaging-software-6073/[/cbtab][cbtab title=”Chicago”]Max Planck Florida Institute For Neuroscience “New Software Automates Brain Imaging.” https://neurosciencenews.com/brain-imaging-software-6073/ (accessed February 6, 2017).[/cbtab][/cbtabs]
Abstract
Automated Remote Focusing, Drift Correction, and Photostimulation to Evaluate Structural Plasticity in Dendritic Spines
Long-term structural plasticity of dendritic spines plays a key role in synaptic plasticity, the cellular basis for learning and memory. The biochemical step is mediated by a complex network of signaling proteins in spines. Two-photon imaging techniques combined with two-photon glutamate uncaging allows researchers to induce and quantify structural plasticity in single dendritic spines. However, this method is laborious and slow, making it unsuitable for high throughput screening of factors necessary for structural plasticity. Here we introduce a MATLAB-based module built for Scanimage to automatically track, image, and stimulate multiple dendritic spines. We implemented an electrically tunable lens in combination with a drift correction algorithm to rapidly and continuously track targeted spines and correct sample movements. With a straightforward user interface to design custom multi-position experiments, we were able to adequately image and produce targeted plasticity in multiple dendritic spines using glutamate uncaging. Our methods are inexpensive, open source, and provides up to a five-fold increase in throughput for quantifying structural plasticity of dendritic spines.
“Automated Remote Focusing, Drift Correction, and Photostimulation to Evaluate Structural Plasticity in Dendritic Spines” by Michael S. Smirnov, Paul R. Evans, Tavita R. Garrett, Long Yan, and Ryohei Yasuda in PLOS ONE. Published online January 23 2017 doi:10.1371/journal.pone.0170586