In this comprehensive review with over 2,450 authors, the topics covered reflect the range of specialist fields within autophagy, and the diversity of animal, plant and fungal cell types that must inevitably invoke autophagy. Even within a single species or organism, the complexity is such that autophagy can be divided into many types.
Autophagy, in general, means ‘biological self-destruction’; it exists in all eukaryotes, including unicellular parasites, plants, fungi and animals. Autophagy is usually considered as a stress response mechanism used under conditions of starvation, infection and diseases such as cancer and neurodegeneration. It is also important in normal development, gut function, wound healing, and (healthy) ageing.
In such cases, autophagy involves the degradation of material in double membrane ‘autophagosomes’ within cells. As the core autophagy process involves several dozen proteins, their precise and context-dependent regulation requires many further proteins and different mechanisms.
The third edition of this Guideline review provides recommendations for best practices, particularly with regard to monitoring and measuring autophagy, and diagnosing/describing the type of autophagy being investigated – multiple assays are currently required and the appropriate choice of these depends upon the question at hand. A major addition to this new version is the presentation of computational tools and resources developed in the last few years to assist autophagy researchers in data analysis and data sharing.
One such resource featured in the review is the Autophagy Regulatory Network database , which provides an integrated and systems-level source for autophagy research in humans.
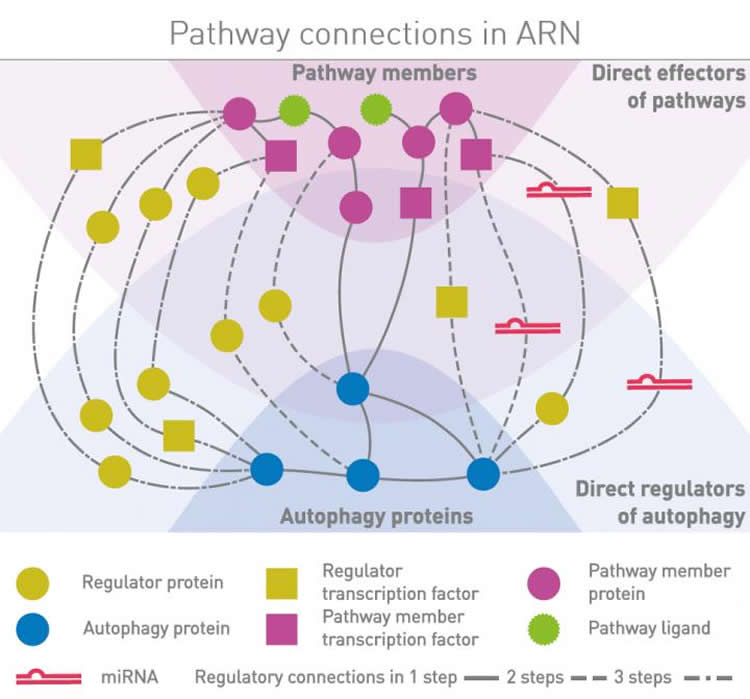
The Autophagy Regulatory Network can be used to analyse autophagy for both global or for gene-specific studies. Currently, it contains data on more than 14,000 proteins, including 38 core autophagy proteins and 113 predicted regulators. A flexible download functionality enables users to customize and filter the database.
Dr Korcsmaros, who contributed the last chapter of the study, said: “We published the Autophagy Regulatory Network exactly one year ago. Since then, the cell biology and autophagy community have started to use it, provided feedbacks and allowed the analysis of autophagy-related systems at a level that was not possible before. Featuring this resource in this key publication (receiving over 2,000 reads just in the first two days), and listing TGAC and IFR among the 2,135 key institutes studying autophagy is a great acknowledgement of our work. We hope that this third edition will serve the community with relevant protocols and tools to study autophagy.”
Funding: TGAC is strategically funded by BBSRC and operates a National Capability to promote the application of genomics and bioinformatics to advance bioscience research and innovation.
Source: Hayley London – The Genome Analysis Center
Image Source: The image is credited to TGAC
Original Research: Full open access research for “Autophagy Regulatory Network — A systems-level bioinformatics resource for studying the mechanism and regulation of autophagy” by Dénes Türei, László Földvári-Nagy, Dávid Fazekas, Dezső Módos, János Kubisch, Tamás Kadlecsik, Amanda Demeter, Katalin Lenti, Péter Csermely, Tibor Vellai and Tamás Korcsmáros in Autophagy. Published online December 2015 doi:10.4161/15548627.2014.994346
Abstract
Autophagy Regulatory Network — A systems-level bioinformatics resource for studying the mechanism and regulation of autophagy
Autophagy is a complex cellular process having multiple roles, depending on tissue, physiological, or pathological conditions. Major post-translational regulators of autophagy are well known, however, they have not yet been collected comprehensively. The precise and context-dependent regulation of autophagy necessitates additional regulators, including transcriptional and post-transcriptional components that are listed in various datasets. Prompted by the lack of systems-level autophagy-related information, we manually collected the literature and integrated external resources to gain a high coverage autophagy database. We developed an online resource, Autophagy Regulatory Network (ARN; http://autophagy-regulation.org), to provide an integrated and systems-level database for autophagy research. ARN contains manually curated, imported, and predicted interactions of autophagy components (1,485 proteins with 4,013 interactions) in humans. We listed 413 transcription factors and 386 miRNAs that could regulate autophagy components or their protein regulators. We also connected the above-mentioned autophagy components and regulators with signaling pathways from the SignaLink 2 resource. The user-friendly website of ARN allows researchers without computational background to search, browse, and download the database. The database can be downloaded in SQL, CSV, BioPAX, SBML, PSI-MI, and in a Cytoscape CYS file formats. ARN has the potential to facilitate the experimental validation of novel autophagy components and regulators. In addition, ARN helps the investigation of transcription factors, miRNAs and signaling pathways implicated in the control of the autophagic pathway. The list of such known and predicted regulators could be important in pharmacological attempts against cancer and neurodegenerative diseases.
“Autophagy Regulatory Network — A systems-level bioinformatics resource for studying the mechanism and regulation of autophagy” by Dénes Türei, László Földvári-Nagy, Dávid Fazekas, Dezső Módos, János Kubisch, Tamás Kadlecsik, Amanda Demeter, Katalin Lenti, Péter Csermely, Tibor Vellai and Tamás Korcsmáros in Autophagy. Published online December 2015 doi:10.4161/15548627.2014.994346