Summary: An international team of researchers have compiled the largest dataset to date that details differences between healthy brains and those with Alzheimer’s disease. The researchers have released the data online and is free to use.
Source: University of Manchester.
A study of the differences between healthy brains and those with Alzheimer’s Disease has produced largest dataset of its type ever.
And the data, developed by a team of researchers led by Dr Richard Unwin at The University of Manchester, is now freely available online for any scientist to use.
The team included researchers from the Universities Manchester, Bristol, Liverpool and Auckland.
The development is an important advance for scientists researching Alzheimer’s.
The team also show that one region of the brain previously thought to be unaffected by the disease, the cerebellum, displayed a series of changes which they think might protect it from damage caused by Alzheimer’s.
The Alzheimer’s Research UK funded study is published in the journal Communications Biology today.
The analysis, mapping the relative levels of over 5,825 distinct proteins across six regions of the brain, generated a massive 24,024 data points.
The brain regions in the study included the more heavily affected Hippocampus, Entorhinal cortex, Cingulate gyrus and the less affected Motor Cortex, Sensory Cortex, and Cerebellum
The samples were donated for research by patients at the New Zealand Brain Bank in Auckland.
Dr Unwin said: “This database provides a huge opportunity for dementia researchers around the world to progress and to follow-up new areas of biology and develop new treatments.
“It could also help validate observations seen in in animal or cell disease models in humans.
“It’s very exciting to be able to make these data public so scientists can access and use this vital information.”
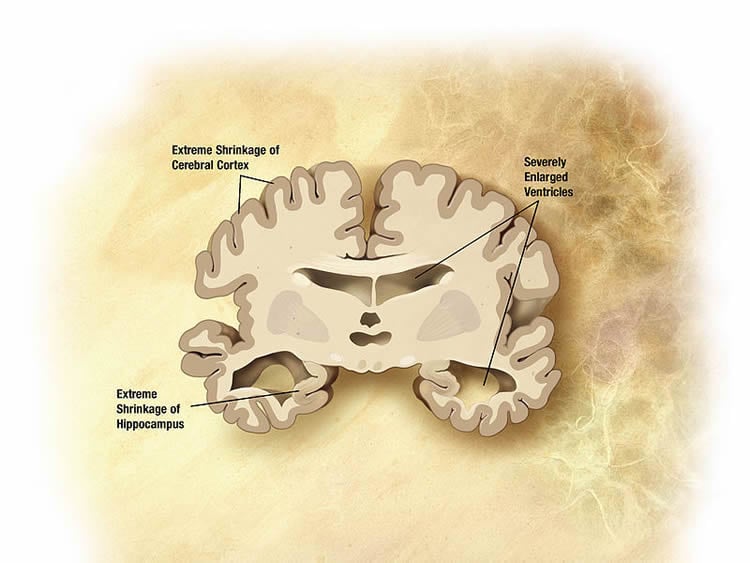
Alzheimer’s Disease arises in the hippocampus and spreads through pathways in the brain.
But by looking at different parts of that pathway, the team were able to observe, for the first time, how Alzheimer’s progresses in more detail.
“We think that the changes we see in the regions affected later on-represent early disease changes, present before cells die,” he said
“These represent good new targets for drug developers, as we know it’s important to try to intervene early.”
In the course of the study, the team hit upon new molecules not previously associated with the disease, representing more targets to develop new drugs.
They also confirm that researchers looking at a range of pathways – including inflammation, Wnt signalling, and metabolic changes in human tissue – are on the right track.
The team identified 129 protein changes which were present in all areas of the brain studied, with at least 44 not previously associated with the disease.
But there were hundreds others which change only in the late-affected regions.
He added: “These new protein changes represent further targets for scientists developing new drugs
“The cerebellum, previously thought be unaffected, displays a significant response at the molecular level.
“Many of the changes here are not seen in other regions and this could imply that this region actively protects itself from disease. We won’t know for sure until we carry out more research.”
Dr Rosa Sancho, Head of Research at Alzheimer’s Research UK, said: “By studying thousands of individual proteins, this exciting research has generated a detailed molecular map of changes that get underway in the brain in Alzheimer’s disease. Making this information freely available online will help researchers to navigate the complex and changing environment of the brain in Alzheimer’s and identify processes that could be targeted by future drugs.
“There are over half a million people in the UK living with Alzheimer’s and there are currently no treatments that can slow or stop the disease from progressing in the brain. Pioneering research like this is driving progress towards new breakthroughs that will change people’s lives.”
Funding: The research was funded by Alzheimer’s Research UK.
Source: Mike Addelman – University of Manchester
Publisher: Organized by NeuroscienceNews.com.
Image Source: NeuroscienceNews.com image is in the public domain.
Original Research: Open access research for “Regional protein expression in human Alzheimer’s brain correlates with disease severity” by Jingshu Xu, Stefano Patassini, Nitin Rustogi, Isabel Riba-Garcia, Benjamin D. Hale, Alexander M Phillips, Henry Waldvogel, Robert Haines, Phil Bradbury, Adam Stevens, Richard L. M. Faull, Andrew W. Dowsey, Garth J. S. Cooper & Richard D. Unwin in Communications Biology. Published February 4 2019.
doi:10.1038/s42003-018-0254-9
[cbtabs][cbtab title=”MLA”]University of Manchester”Biggest Ever Map of Human Alzheimer’s Brain Published.” NeuroscienceNews. NeuroscienceNews, 44 February 2019.
<https://neurosciencenews.com/alzheimers-brain-map-10682/>.[/cbtab][cbtab title=”APA”]University of Manchester(2019, February 44). Biggest Ever Map of Human Alzheimer’s Brain Published. NeuroscienceNews. Retrieved February 44, 2019 from https://neurosciencenews.com/alzheimers-brain-map-10682/[/cbtab][cbtab title=”Chicago”]University of Manchester”Biggest Ever Map of Human Alzheimer’s Brain Published.” https://neurosciencenews.com/alzheimers-brain-map-10682/ (accessed February 44, 2019).[/cbtab][/cbtabs]
Abstract
Regional protein expression in human Alzheimer’s brain correlates with disease severity
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that currently affects 36 million people worldwide with no effective treatment available. Development of AD follows a distinctive pattern in the brain and is poorly modelled in animals. Therefore, it is vital to widen the spatial scope of the study of AD and prioritise the study of human brains. Here we show that functionally distinct human brain regions display varying and region-specific changes in protein expression. These changes provide insights into the progression of disease, novel AD-related pathways, the presence of a gradient of protein expression change from less to more affected regions and a possibly protective protein expression profile in the cerebellum. This spatial proteomics analysis provides a framework which can underpin current research and open new avenues to enhance molecular understanding of AD pathophysiology, provide new targets for intervention and broaden the conceptual frameworks for future AD research.