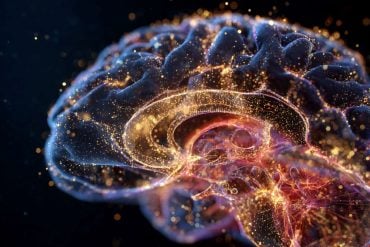
Summary: Researchers have developed TRISCO, a cutting-edge microscopy method that enables 3D RNA analysis in whole, intact mouse brains without slicing them into sections. TRISCO reveals RNA molecules’ spatial distribution, providing unprecedented insights into the brain’s complex structure and function.
The technique has the potential to analyze up to 100 RNA molecules simultaneously, paving the way for advanced studies on brain function and disease. This breakthrough could revolutionize our understanding of neurological disorders and lead to innovative treatments.
Key Facts:
- TRISCO allows three-dimensional RNA imaging in whole, intact mouse brains.
- It can analyze multiple RNA molecules simultaneously, soon expanding to 100.
- The method is applicable to larger brains and other tissues like heart and kidney.
Source: Karolinska Institute
Researchers at Karolinska Institutet and Karolinska University Hospital have developed a groundbreaking microscopy method that enables detailed three-dimensional (3D) RNA analysis at cellular resolution in whole intact mouse brains.
The new method, called TRISCO, has the potential to transform our understanding of brain function, both in normal conditions and in disease, according to the new study published in Science.
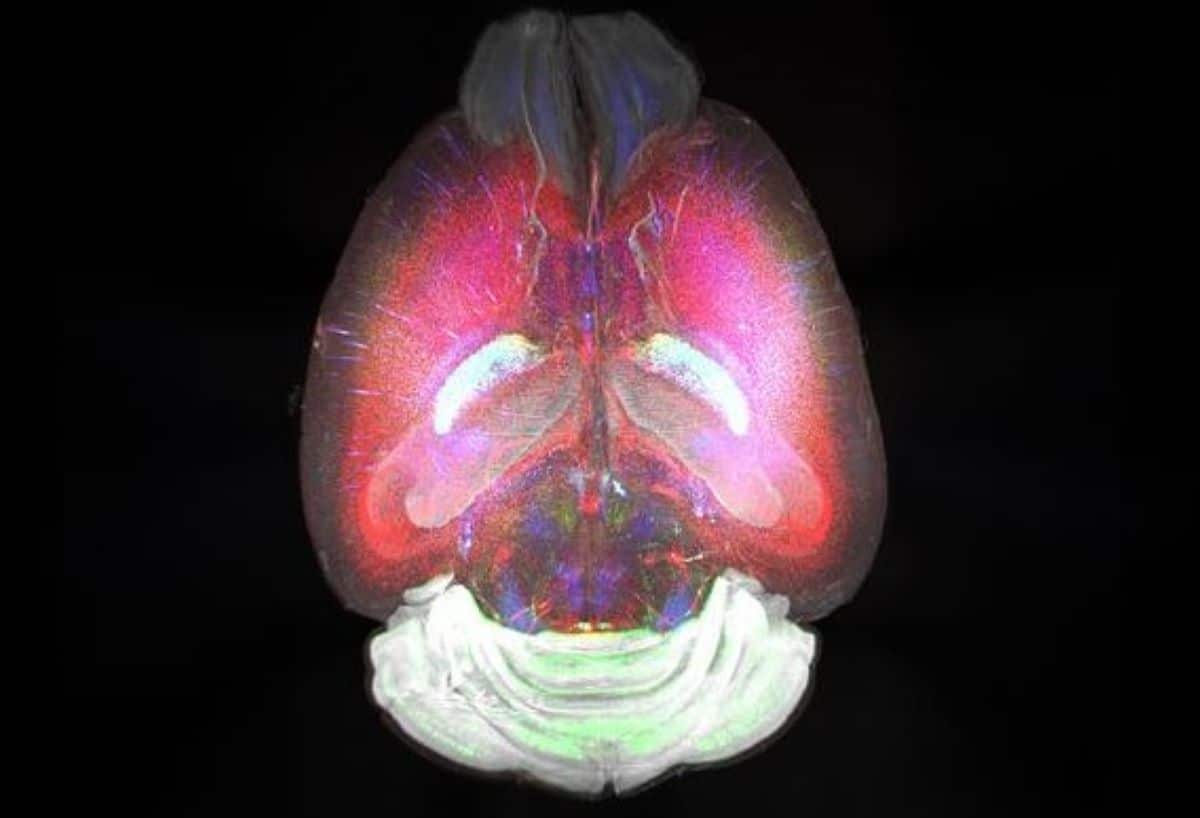
Despite great advances in RNA analysis, linking RNA data to its spatial context has long been a challenge, especially in intact 3D tissue volumes. The TRISCO method now makes it possible to perform three-dimensional RNA imaging of whole mouse brains without the need to slice the brain into thin sections, which was previously necessary.
“This method is a powerful tool that can drive brain research forward. With TRISCO, we can study the complex anatomical structure of the brain in a way that was previously not possible,” says Per Uhlén, professor at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet, and the study’s last author.
In the study, up to three different RNA molecules were analysed simultaneously. The next step for the researchers is to expand the number of RNA molecules that can be studied to around a hundred, using a technique called multiplex RNA analysis. This could provide even more detailed information about brain function and disease states.
The TRISCO approach opens up new possibilities to understand the complexity of the brain in depth, which in turn can lead to the development of new treatments for various brain diseases.
“We look forward to continuing our research and exploring the many possibilities offered by this new technique,” says Shigeaki Kanatani, a research specialist in Uhlén’s laboratory and the first author of the study.
Not only is TRISCO suitable for studying intact mouse brains, but the study demonstrates it can be used for larger brains, such as those of guinea pigs, and various tissues like kidney, heart, and lung. The study is a collaboration between Karolinska Institutet and Karolinska University Hospital.
“Our laboratory has several collaborations with clinically active researchers at Karolinska University Hospital. It is crucial for biomedical research that basic researchers and clinicians collaborate and understand each other,” says Per Uhlén.
Funding: The study has been funded by the Swedish Research Council, Swedish Brain Foundation and Swedish Cancer Society. Some of the co-authors are employed and own shares in the Danish company Gubra. No other conflicts of interest are reported.
About this RNA and neuroimaging research news
Author: Press Office
Source: Karolinska Institute
Contact: Press Office – Karolinska Institute
Image: The image is credited to Science
Original Research: Closed access.
“Whole-Brain Spatial Transcriptional Analysis at Cellular Resolution” by Per Uhlén et al. Science
Abstract
Whole-Brain Spatial Transcriptional Analysis at Cellular Resolution
Recent advances in RNA analysis have deepened our understanding of cellular states in biological tissues. However, a substantial gap remains in integrating RNA expression data with spatial context across organs, primarily owing to the challenges associated with RNA detection within intact tissue volumes.
Here, we developed Tris buffer–mediated retention of in situ hybridization chain reaction signal in cleared organs (TRISCO), an effective tissue-clearing method designed for whole-brain spatial three-dimensional (3D) RNA imaging.
TRISCO resolved several crucial issues, including the preservation of RNA integrity, achieving uniform RNA labeling, and enhancing tissue transparency.
We tested TRISCO using a broad range of cell-identity markers, noncoding and activity-dependent RNAs, within diverse organs of varying sizes and species.
TRISCO thus emerges as a powerful tool for single-cell, whole-brain, 3D imaging that enables comprehensive transcriptional spatial analysis across the entire brain.