Summary: Deep convolutional neural networks were used to help process neonatal brain image data. Modified LiviaNET and HyperDense-Net deep learning neural networks are discussed.
Source: University of Montreal
Canadian scientists have developed an innovative new technique that uses artificial intelligence to better define the different sections of the brain in newborns during a magnetic resonance imaging (MRI) exam.
The results of this study — a collaboration between researchers at Montreal’s CHU Sainte-Justine children’s hospital and the ÉTS engineering school — are published today in Frontiers in Neuroscience.
“This is one of the first times that artificial intelligence has been used to better define the different parts of a newborn’s brain on an MRI: namely the grey matter, white matter and cerebrospinal fluid,” said Dr. Gregory A. Lodygensky, a neonatologist at CHU Sainte-Justine and professor at Université de Montréal.
“Until today, the tools available were complex, often intermingled and difficult to access,” he added.
In collaboration with Professor Jose Dolz, an expert in medical image analysis and machine learning at ÉTS, the researchers were able to adapt the tools to the specificities of the neonatal setting and then validate them.
This new technique allows babies’ brains to be examined quickly, accurately and reliably. Scientists see it as a major asset for supporting research that not only addresses brain development in neonatal care, but also the effectiveness of neuroprotective strategies.
In evaluating a range of tools available in artificial intelligence, CHU Sainte-Justine researchers found that these tools had limitations, particularly with respect to pediatric research. Today’s neuroimaging analysis programs are primarily designed to work on “adult” MRIs. The cerebral immaturity of newborns, with an inversion of the contrasts between grey matter and white matter, complicates such analyses.
Inspired by Dolz’s most recent work, the researchers proposed an artificial neural network that learns how to efficiently combine information from several MRI sequences. This methodology made it possible to better define the different parts of the brain in the newborn automatically and to establish a new benchmark for this problem.
“We’ve decided not only to share the results of our study on open source, but also the computer code, so that brain researchers everywhere can take advantage of it, all of which benefits patients,” said Dolz.
CHU Sainte-Justine is one of the most important players in the Canadian Neonatal Brain Platform and also has one of the largest neonatal units in Canada specializing in neurodevelopment. As part of the platform, research teams are implementing projects like this one with the aim of improving the long-term health of those newborns who are most vulnerable to brain injury.
“In studies to assess the positive and negative impact of different therapies on the maturation of babies’ brains, we need to have the ability to quantify brain structures with certainty and reliability,” Lodygensky said. “By offering the scientific community the fruits of all our discoveries, we are helping them, while generating an extraordinary benefit for at-risk newborns.”
He added: “We now want to democratize this tool so that it becomes the benchmark for the study of brain structure in newborns around the world. To this end, we are continuing to work on its generalizability — that is, its use on MRI data acquired in different hospitals.”
Source:
University of Montreal
Media Contact:
Florence Meney
Image:
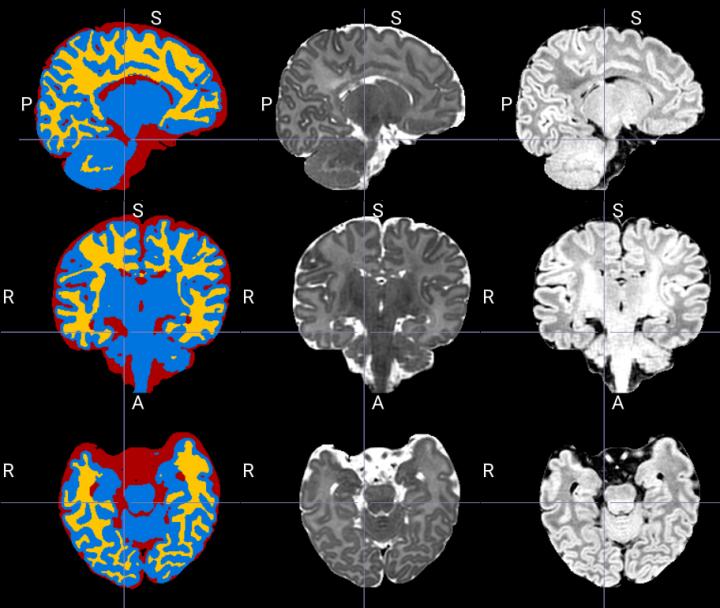
Example of segmentation produced by the tool which separates the structures in cerebrospinal fluid (red), grey matter (blue) and white matter (yellow) from MRI images T2 (middle column) and T1 (right column).
Credit:
CHU Sainte-Justine
Original Research: Open Access
“Using deep convolutional neural networks for neonatal brain image segmentation” was published March 26, 2020 in Frontiers in Neuroscience. The first author is Yang Ding, PhD, under the direction of Gregory A. Lodygensky. The primary authors are Gregory A. Lodygensky, MD, Clinical Associate Professor in the Department of Pediatrics at Université de Montréal and Clinician-Researcher at CHU Sainte-Justine, and Jose Dolz, PhD, Assistant Professor in the Department of Software Engineering and Information Technology at the École de technologie supérieure (ÉTS). The study was supported by the Brain Canada Foundation.
http://dx.doi.org/10.3389/fnins.2020.00207
“Using Deep Convolutional Neural Networks for Neonatal Brain Image Segmentation”
Yang Ding, Rolando Acosta, Vicente Enguix, Sabrina Suffren, Janosch Ortmann, David Luck, Jose Dolz and Gregory A. Lodygensky
Frontiers in Neuroscience
Introduction: Deep learning neural networks are especially potent at dealing with structured data, such as images and volumes. Both modified LiviaNET and HyperDense-Net performed well at a prior competition segmenting 6-month-old infant magnetic resonance images, but neonatal cerebral tissue type identification is challenging given its uniquely inverted tissue contrasts. The current study aims to evaluate the two architectures to segment neonatal brain tissue types at term equivalent age.
Methods: Both networks were retrained over 24 pairs of neonatal T1 and T2 data from the Developing Human Connectome Project public data set and validated on another eight pairs against ground truth. We then reported the best-performing model from training and its performance by computing the Dice similarity coefficient (DSC) for each tissue type against eight test subjects.
Results: During the testing phase, among the segmentation approaches tested, the dual-modality HyperDense-Net achieved the best statistically significantly test mean DSC values, obtaining 0.94/0.95/0.92 for the tissue types and took 80 h to train and 10 min to segment, including preprocessing. The single-modality LiviaNET was better at processing T2-weighted images than processing T1-weighted images across all tissue types, achieving mean DSC values of 0.90/0.90/0.88 for gray matter, white matter, and cerebrospinal fluid, respectively, while requiring 30 h to train and 8 min to segment each brain, including preprocessing.
Discussion: Our evaluation demonstrates that both neural networks can segment neonatal brains, achieving previously reported performance. Both networks will be continuously retrained over an increasingly larger repertoire of neonatal brain data and be made available through the Canadian Neonatal Brain Platform to better serve the neonatal brain imaging research community.
About the CHU Sainte-Justine Research Center
The CHU Sainte-Justine Research Center is a leading mother-child research institution affiliated with Université de Montréal. It brings together more than 210 research investigators, including over 110 clinician-scientists, as well as 450 graduate and postgraduate students focused on finding innovative prevention means, faster and less invasive treatments, as well as personalized approaches to medicine. The Center is part of CHU Sainte-Justine, which is the largest mother-child center in Canada. For more information, go to research.chusj.org.
About ÉTS
ÉTS is one of the ten constituents of the University of Québec network. It trains engineers and researchers recognized for their practical and innovative approach, the development of new technologies and their ability to transfer their knowledge to private enterprise. CSRankings places ÉTS in the vanguard of the artificial vision branch of artificial intelligence: it ranks first in Québec and sixth in Canada for scientific publications in this domain. For more information, visit: etsmtl.ca.