Summary: Significantly more DNA sequences repeat in people with schizophrenia than in control individuals, a new study reports.
Source: Nagoya University.
International researchers centered at Nagoya University use a highly sensitive technique to identify significantly more DNA sequence repeats in patients with schizophrenia than in control individuals, and outline a possible link between genome instability and disease.
Variations in the number of DNA sequence repeats are known to exist between individuals. Some of these copy number variations (CNVs) are associated with disease, as with schizophrenia, wherein rare CNVs on chromosomes, including 1, 15, 16, and 22, are more common among patients than among controls. However, previous studies have not fully investigated the effect of particular CNVs, especially those on the X chromosome, on patient characteristics. Nor have they examined the genes responsible for CNVs in schizophrenia, which would aid understanding of disease development.
An international research team centered at Nagoya University has now used a technique based on DNA fragments labeled with different fluorescent markers to reveal high levels of genetic heterogeneity (whereby several different genetic defects can cause the same symptoms) in schizophrenia. They also observed that the CNVs associated with schizophrenia affected gene categories controlling repair of DNA damage, which may underlie disease mechanisms. The study was reported in the Springer Nature journal Molecular Psychiatry.
The technique the researchers used was more sensitive and of a higher resolution than previous methods. This allowed detection of more CNVs, especially small ones, which accounted for 70% of all CNVs. Significantly more clinically important CNVs were seen in the DNA of 1,699 patients with schizophrenia than in the 824 control individuals, while abnormal numbers of X chromosomes were also associated with disease. High genetic heterogeneity was revealed by the detection of these CNVs at 67 different regions in 9% of patients. Further variation was observed in the effect of the CNVs on patient characteristics in that some CNVs were carried by controls without causing any symptoms.
“Patients with clinically important CNVs showed a range of characteristics, such as developmental problems and refusal to accept treatment,” first author Itaru Kushima says. “And, the presence of two CNVs resulted in a more severe phenotype.”
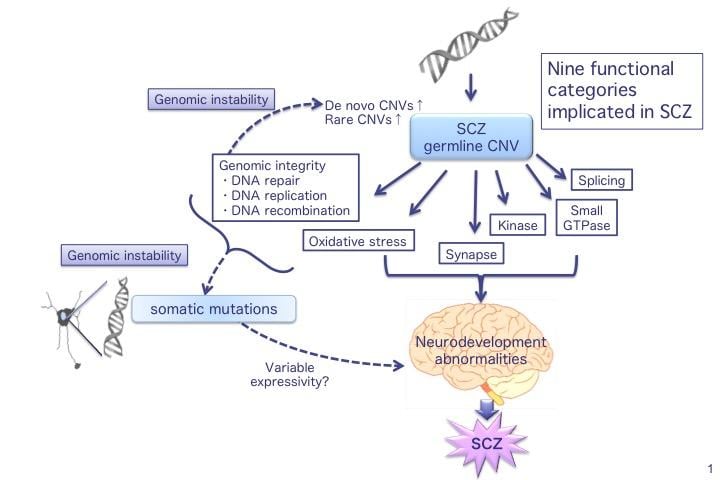
The researchers looked closely at the genetic regions containing CNVs to identify several gene categories associated with schizophrenia that may be affected by genetic disturbance. These include oxidative stress response, which leads to DNA damage when imbalanced, and genomic integrity, involving DNA repair and replication.
“We propose that CNVs affecting oxidative stress response and genomic integrity lead to genomic instability that may cause further CNVs”, corresponding author Norio Ozaki says. “This model helps explain the new CNVs seen in previous studies on schizophrenia, as well as the differences in affected patients’ phenotypes.”
Funding: Funding provided by Japan Agency for Medical Research and Development, Ministry of Education, Culture, Sports, Science & Technology in Japan.
Source: Koomi Sung – Nagoya University
Image Source: This NeuroscienceNews.com image is credited to Itaru Kushima and Norio Ozaki.
Original Research: Abstract for “High-resolution copy number variation analysis of schizophrenia in Japan” by I Kushima, B Aleksic, M Nakatochi, T Shimamura, T Shiino, A Yoshimi, H Kimura, Y Takasaki, C Wang, J Xing, K Ishizuka, T Oya-Ito, Y Nakamura, Y Arioka, T Maeda, M Yamamoto, M Yoshida, H Noma, S Hamada, M Morikawa, Y Uno, T Okada, T Iidaka, S Iritani, T Yamamoto, M Miyashita, A Kobori, M Arai, M Itokawa, M -C Cheng, Y -A Chuang, C -H Chen, M Suzuki, T Takahashi, R Hashimoto, H Yamamori, Y Yasuda, Y Watanabe, A Nunokawa, T Someya, M Ikeda, T Toyota, T Yoshikawa, S Numata, T Ohmori, S Kunimoto, D Mori, N Iwata and N Ozaki in Molecular Psychiatry. Published online May 31 2016 doi:10.1038/mp.2016.88
[cbtabs][cbtab title=”MLA”]Nagoya University. “DNA Instability in Schizophrenia.” NeuroscienceNews. NeuroscienceNews, 24 June 2016.
<https://neurosciencenews.com/synapse-neural-communication-4556/>.[/cbtab][cbtab title=”APA”]Nagoya University. (2016, June 24). DNA Instability in Schizophrenia. NeuroscienceNew. Retrieved June 24, 2016 from https://neurosciencenews.com/synapse-neural-communication-4556/[/cbtab][cbtab title=”Chicago”]Nagoya University. “DNA Instability in Schizophrenia.” https://neurosciencenews.com/synapse-neural-communication-4556/ (accessed June 24, 2016).[/cbtab][/cbtabs]
Abstract
High-resolution copy number variation analysis of schizophrenia in Japan
Recent schizophrenia (SCZ) studies have reported an increased burden of de novo copy number variants (CNVs) and identified specific high-risk CNVs, although with variable phenotype expressivity. However, the pathogenesis of SCZ has not been fully elucidated. Using array comparative genomic hybridization, we performed a high-resolution genome-wide CNV analysis on a mainly (92%) Japanese population (1699 SCZ cases and 824 controls) and identified 7066 rare CNVs, 70.0% of which were small (<100 kb). Clinically significant CNVs were significantly more frequent in cases than in controls (odds ratio=3.04, P=9.3 × 10−9, 9.0% of cases). We confirmed a significant association of X-chromosome aneuploidies with SCZ and identified 11 de novo CNVs (e.g., MBD5 deletion) in cases. In patients with clinically significant CNVs, 41.7% had a history of congenital/developmental phenotypes, and the rate of treatment resistance was significantly higher (odds ratio=2.79, P=0.0036). We found more severe clinical manifestations in patients with two clinically significant CNVs. Gene set analysis replicated previous findings (e.g., synapse, calcium signaling) and identified novel biological pathways including oxidative stress response, genomic integrity, kinase and small GTPase signaling. Furthermore, involvement of multiple SCZ candidate genes and biological pathways in the pathogenesis of SCZ was suggested in established SCZ-associated CNV loci. Our study shows the high genetic heterogeneity of SCZ and its clinical features and raises the possibility that genomic instability is involved in its pathogenesis, which may be related to the increased burden of de novo CNVs and variable expressivity of CNVs.
“High-resolution copy number variation analysis of schizophrenia in Japan” by I Kushima, B Aleksic, M Nakatochi, T Shimamura, T Shiino, A Yoshimi, H Kimura, Y Takasaki, C Wang, J Xing, K Ishizuka, T Oya-Ito, Y Nakamura, Y Arioka, T Maeda, M Yamamoto, M Yoshida, H Noma, S Hamada, M Morikawa, Y Uno, T Okada, T Iidaka, S Iritani, T Yamamoto, M Miyashita, A Kobori, M Arai, M Itokawa, M -C Cheng, Y -A Chuang, C -H Chen, M Suzuki, T Takahashi, R Hashimoto, H Yamamori, Y Yasuda, Y Watanabe, A Nunokawa, T Someya, M Ikeda, T Toyota, T Yoshikawa, S Numata, T Ohmori, S Kunimoto, D Mori, N Iwata and N Ozaki in Molecular Psychiatry. Published online May 31 2016 doi:10.1038/mp.2016.88