New large-scale computational analysis of gene expression in single cells in the brain identifies distinct cell types.
Understanding the cellular building blocks of the brain, including the number and diversity of cell types, is a fundamental step toward understanding brain function. Researchers at the Allen Institute for Brain Science have created a detailed taxonomy of cells in the mouse visual cortex based on single-cell gene expression, identifying 49 distinct cell types in the largest collection of individual adult cortical neurons characterized by gene expression published to date. The work appears this month online in Nature Neuroscience.
“Studying any system requires knowing what the system is made of,” says Bosiljka Tasic, Ph.D., Assistant Investigator at the Allen Institute for Brain Science. “There are many ways to define the brain’s cellular building blocks. Our approach was to look at all the genes that are expressed in individual cells in the mouse visual cortex and use that information to classify the cells.”
The team developed a technique to isolate single cells from the adult mouse brain, and then obtained genome-wide gene expression data from these individual cells. Each cell expresses thousands of genes, making the cell classification problem an enormous computational task.
“Initially, the problem of classifying cells is like sorting Skittles in the dark,” says Vilas Menon, Scientist II at the Allen Institute for Brain Science. “With single-cell gene expression data, we get the equivalent of color, or type, information, but we still have to extract it from the large-scale data set. Ultimately, we wanted to figure out how many types there were in an unbiased, data-driven way.”
Tasic, Menon and their team used computational dimension reduction techniques, which collapse genes with similar expression patterns into gene sets. When single cells were analyzed by clustering in this lower-dimensional space, 49 distinct groups appeared based on unique combinations of genes they express, including 42 neuronal cell types and 7 non-neuronal types.
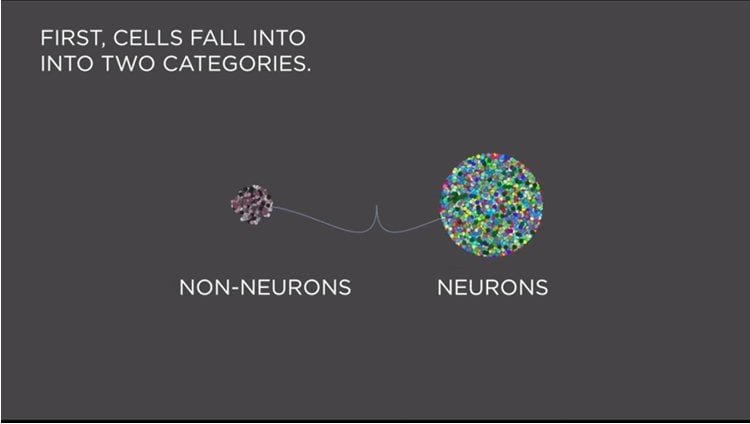
“Our human cortex is what gives rise to our unique thoughts and perceptions,” says Christof Koch, Ph.D., President and Chief Scientific Officer at the Allen Institute for Brain Science. “Having this kind of objective analysis of cell types in this region of the brain is a basic piece of understanding we need, and provides a baseline for looking at other regions of the mouse brain and also at the human brain.”
The data from this single cell analysis approach agree with and complement the Allen Brain Atlas: a brain-wide gene expression atlas of the mouse brain.
“Our unit of analysis was a single cell and all genes within each cell, but in our process, we lost fine spatial information,” says Tasic. “But then, we were able to use our Allen Brain Atlas, which has brain-wide analysis of each gene, one gene at a time at cellular resolution, to more precisely locate each cell type. Our work is one more step toward assigning genes to specific cell types and then helping investigate what these genes do, how they work together, and how they ultimately make our nervous systems and us who we are.”
The mammalian brain is composed of various cell populations that differ based on their molecular, morphological, electrophysiological and functional characteristics. Classifying these cells into types is one of the essential approaches to defining the diversity of brain’s building blocks. We created a cellular taxonomy of the mouse primary visual cortex by analyzing gene expression patterns, at the single cell level, of more than 1,600 cells. Using transgenic mice, we isolated fluorescently labeled cells from their brains and then sequenced the transcriptomes of individual cells. To identify the different cell types, we employed an iterative unbiased classification method (cluster analysis) that examined all expressed genes and was blind to the origin of cells. Credit: Allen Institute.
“Categorizing the cells in visual cortex into these distinct types that are marked by specific genes will enable us to begin to understand what these cells and types do in the brain,” says Hongkui Zeng, Ph.D., Investigator of Cell and Circuit Genetics at the Allen Institute for Brain Science. “Next, we can investigate how gene expression correlates with the anatomical, physiological and functional properties of the cells, how these cell types are connected with each other, and how they work together to process and make sense of the visual information the brain receives from the outside world. This will ultimately shed light on the inner workings of the brain.”
Source: Rob Piercy – Allen Institute
Image Credit: The image is adapted from the Allen Institute video
Video Source: The video is credited to Allen Institute
Original Research: Abstract for “Adult mouse cortical cell taxonomy revealed by single cell transcriptomics” by Bosiljka Tasic, Vilas Menon, Thuc Nghi Nguyen, Tae Kyung Kim, Tim Jarsky, Zizhen Yao, Boaz Levi, Lucas T Gray, Staci A Sorensen, Tim Dolbeare, Darren Bertagnolli, Jeff Goldy, Nadiya Shapovalova, Sheana Parry, Changkyu Lee, Kimberly Smith, Amy Bernard, Linda Madisen, Susan M Sunkin, Michael Hawrylycz, Christof Koch and Hongkui Zeng in Nature Neuroscience. Published online January 4 2016 doi:10.1038/nn.4216
Abstract
Adult mouse cortical cell taxonomy revealed by single cell transcriptomics
Nervous systems are composed of various cell types, but the extent of cell type diversity is poorly understood. We constructed a cellular taxonomy of one cortical region, primary visual cortex, in adult mice on the basis of single-cell RNA sequencing. We identified 49 transcriptomic cell types, including 23 GABAergic, 19 glutamatergic and 7 non-neuronal types. We also analyzed cell type–specific mRNA processing and characterized genetic access to these transcriptomic types by many transgenic Cre lines. Finally, we found that some of our transcriptomic cell types displayed specific and differential electrophysiological and axon projection properties, thereby confirming that the single-cell transcriptomic signatures can be associated with specific cellular properties.
“Adult mouse cortical cell taxonomy revealed by single cell transcriptomics” by Bosiljka Tasic, Vilas Menon, Thuc Nghi Nguyen, Tae Kyung Kim, Tim Jarsky, Zizhen Yao, Boaz Levi, Lucas T Gray, Staci A Sorensen, Tim Dolbeare, Darren Bertagnolli, Jeff Goldy, Nadiya Shapovalova, Sheana Parry, Changkyu Lee, Kimberly Smith, Amy Bernard, Linda Madisen, Susan M Sunkin, Michael Hawrylycz, Christof Koch and Hongkui Zeng in Nature Neuroscience. Published online January 4 2016 doi:10.1038/nn.4216