Summary: According to a new study, a computer program was almost twice as accurate as neuroradiologists in determining whether abnormal brain tissue seen on an MRI scan was due to radiation or brain cancer.
Source: Case Western Reserve.
Case Western Reserve University-led research could speed identification of recurrent tumors, eliminate costly and risky brain biopsies.
Computer programs have defeated humans in Jeopardy!, chess and Go. Now a program developed at Case Western Reserve University has outperformed physicians on a more serious matter.
The program was nearly twice as accurate as two neuroradiologists in determining whether abnormal tissue seen on magnetic resonance images (MRI) were dead brain cells caused by radiation, called radiation necrosis, or if brain cancer had returned.
The direct comparison is part of a feasibility study published in the American Journal of Neuroradiology today.
“One of the biggest challenges with the evaluation of brain tumor treatment is distinguishing between the confounding effects of radiation and cancer recurrence,” said Pallavi Tiwari, assistant professor of biomedical engineering at Case Western Reserve and leader of the study. “On an MRI, they look very similar.”
But treatments for radiation necrosis and cancer recurrence are far different. Quick identification can help speed prognosis, therapy and improve patient outcomes, the researchers say.
With further confirmation of its accuracy, radiologists using their expertise and the program may eliminate unnecessary and costly biopsies Tiwari said. Brain biopsies are currently the only definitive test but are highly invasive and risky, causing considerable morbidity and mortality.
To develop the program, the researchers employed machine learning algorithms in conjunction with radiomics, the term used for features extracted from images using computer algorithms. The engineers, scientists and physicians trained the computer to identify radiomic features that discriminate between brain cancer and radiation necrosis, using routine follow-up MRI scans from 43 patients. The images were all from University Hospitals Case Medical Center.
The team then developed algorithms to find the most discriminating radiomic features, in this case, textures that can’t be seen by simply eyeballing the images.
“What the algorithms see that the radiologists don’t are the subtle differences in quantitative measurements of tumor heterogeneity and breakdown in microarchitecture on MRI, which are higher for tumor recurrence,” said Tiwari, who was appointed to the Department of Biomedical Engineering by the Case Western Reserve School of Medicine.
More specifically, while the physicians use the intensity of pixels on MRI scans as a guide, the computer looks at the edges of each pixel, explained Anant Madabhushi, F. Alex Nason professor II of biomedical engineering at Case Western Reserve, and study co-author.
“If the edges all point to the same direction, the architecture is preserved,” said Madabhushi, who also directs the Center of Computational Imaging and Personalized Diagnostics at CWRU. “If they point in different directions, the architecture is disrupted—the entropy, or disorder, and heterogeneity are higher. “
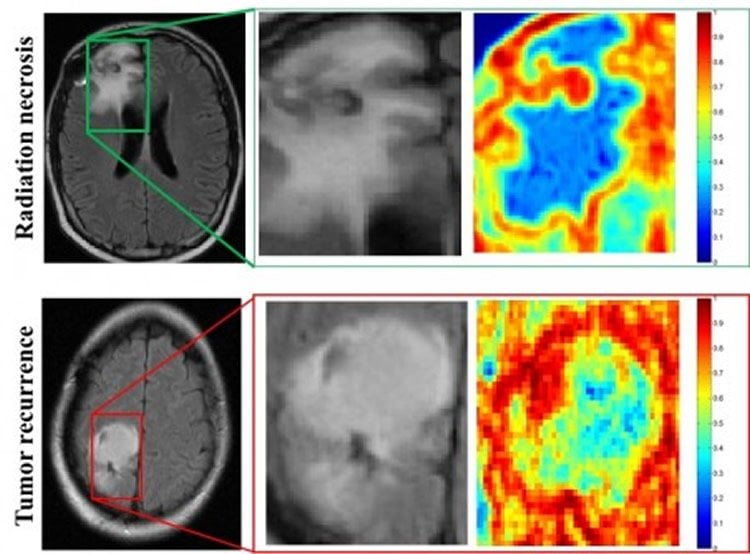
In the direct comparison, two physicians and the computer program analyzed MRI scans from 15 patients from University of Texas Southwest Medical Center. One neuroradiologist diagnosed seven patients correctly, and the second physician correctly diagnosed eight patients. The computer program was correct on 12 of the 15.
Tiwari and Madabhushi don’t expect the computer program would be used alone, but as a decision support to assist neuroradiologists in improving their confidence in identifying a suspicious lesion as radiation necrosis or cancer recurrence.
Next, the researchers are seeking to validate and the algorithms’ accuracy using a much larger collection of images from across different sites.
Tiwari and Madabhushi worked with Case School of Engineering’s Prateek Prasanna, PhD student, and Gagandeep Singh, MBBS, medical research fellow; Case Western Reserve School of Medicine’s Leo Wolansky, MD, professor of radiology; Mark Cohen, MD, professor of pathology; Ameya Nayate, assistant professor of radiology; Amit Gupta, Fellow, Nuclear Radiology, University Hospitals Case Medical Center; Andrew Sloan, MD, Peter D. Cristal Chair of Neurosurgical Oncology; and Lisa Rogers, DO, professor of neurology; and University of Texas Southwest Medical Center’s Marco Pinho, MD, assistant professor of radiology; and Kimmo Hattanpaa, MD, PhD associate professor of pathology.
Funding: Research reported in this publication was supported by the Coulter Translational Award, National Cancer Institute of the National Institutes of Health under award numbers R01CA136535-01, R01CA140772-01, and R21CA167811-01; the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under award number R43EB015199-01; the National Science Foundation under award number IIP-1248316; the Clinical and Translational Science Collaborative (CTSC) Award UL1TR 000439, and the SOURCE Summer Research Program and the Case Western Alumni Association.
Source: Kevin Mayhood – Case Western Reserve
Image Source: NeuroscienceNews.com image is credited to Pallavi Tiwari.
Original Research: Abstract for “Computer-Extracted Texture Features to Distinguish Cerebral Radionecrosis from Recurrent Brain Tumors on Multiparametric MRI: A Feasibility Study” by P. Tiwari, P. Prasanna, L. Wolansky, M. Pinho, M. Cohen, A.P. Nayate, A. Gupta, G. Singh, K. Hattanpaa, A. Sloan, L. Rogers and A. Madabhushi in American Journal of Neuroradiology. Published online September 15 2016 doi:10.3174/ajnr.A4931
Title Correction: We thank Mark Weatherall for suggesting a more accurate title. This post was previously titled Computer Program Beats Doctors at Brain Cancer Diagnosis which didn’t reflect the article content effectively.
[cbtabs][cbtab title=”MLA”]Case Western Reserve. “Computer Program Beats Doctors at Distinguishing Brain Tumors from Radiation Changes.” NeuroscienceNews. NeuroscienceNews, 16 September 2016.
<https://neurosciencenews.com/ai-brain-cancer-neurology-5058/>.[/cbtab][cbtab title=”APA”]Case Western Reserve. (2016, September 16). Computer Program Beats Doctors at Distinguishing Brain Tumors from Radiation Changes. NeuroscienceNews. Retrieved September 16, 2016 from https://neurosciencenews.com/ai-brain-cancer-neurology-5058/[/cbtab][cbtab title=”Chicago”]Case Western Reserve. “Computer Program Beats Doctors at Distinguishing Brain Tumors from Radiation Changes.” https://neurosciencenews.com/ai-brain-cancer-neurology-5058/ (accessed September 16, 2016).[/cbtab][/cbtabs]
Abstract
An Extension of the Procedural Deficit Hypothesis from Developmental Language Disorders to Mathematical Disability
BACKGROUND AND PURPOSE: Despite availability of advanced imaging, distinguishing radiation necrosis from recurrent brain tumors noninvasively is a big challenge in neuro-oncology. Our aim was to determine the feasibility of radiomic (computer-extracted texture) features in differentiating radiation necrosis from recurrent brain tumors on routine MR imaging (gadolinium T1WI, T2WI, FLAIR).
MATERIALS AND METHODS: A retrospective study of brain tumor MR imaging performed 9 months (or later) post-radiochemotherapy was performed from 2 institutions. Fifty-eight patient studies were analyzed, consisting of a training (n = 43) cohort from one institution and an independent test (n = 15) cohort from another, with surgical histologic findings confirmed by an experienced neuropathologist at the respective institutions. Brain lesions on MR imaging were manually annotated by an expert neuroradiologist. A set of radiomic features was extracted for every lesion on each MR imaging sequence: gadolinium T1WI, T2WI, and FLAIR. Feature selection was used to identify the top 5 most discriminating features for every MR imaging sequence on the training cohort. These features were then evaluated on the test cohort by a support vector machine classifier. The classification performance was compared against diagnostic reads by 2 expert neuroradiologists who had access to the same MR imaging sequences (gadolinium T1WI, T2WI, and FLAIR) as the classifier.
RESULTS: On the training cohort, the area under the receiver operating characteristic curve was highest for FLAIR with 0.79; 95% CI, 0.77–0.81 for primary (n = 22); and 0.79, 95% CI, 0.75–0.83 for metastatic subgroups (n = 21). Of the 15 studies in the holdout cohort, the support vector machine classifier identified 12 of 15 studies correctly, while neuroradiologist 1 diagnosed 7 of 15 and neuroradiologist 2 diagnosed 8 of 15 studies correctly, respectively.
CONCLUSIONS: Our preliminary results suggest that radiomic features may provide complementary diagnostic information on routine MR imaging sequences that may improve the distinction of radiation necrosis from recurrence for both primary and metastatic brain tumors.
“Computer-Extracted Texture Features to Distinguish Cerebral Radionecrosis from Recurrent Brain Tumors on Multiparametric MRI: A Feasibility Study” by P. Tiwari, P. Prasanna, L. Wolansky, M. Pinho, M. Cohen, A.P. Nayate, A. Gupta, G. Singh, K. Hattanpaa, A. Sloan, L. Rogers and A. Madabhushi in American Journal of Neuroradiology. Published online September 15 2016 doi:10.3174/ajnr.A4931