Summary: A new study introduces BARseq—a rapid, cost-effective method for mapping brain cells, revealing new insights into how our brains are structured at a cellular level. Researchers used BARseq to classify millions of neurons across multiple mouse brains, discovering unique ‘cellular signatures’ that define each brain region.
The study also highlighted how sensory deprivation, such as loss of sight, can significantly reorganize these neuronal structures, underscoring the importance of sensory experiences in shaping the brain. This new tool not only advances our understanding of brain architecture but also opens up possibilities for exploring brain changes associated with diseases.
Key Facts:
- BARseq technology enables rapid and extensive mapping of neurons across the brain, identifying distinct cellular signatures unique to each brain region.
- Sensory experiences, particularly vision, play a critical role in maintaining and shaping the distinct cellular identities of different brain areas.
- The BARseq method is both more affordable and faster than previous brain mapping technologies, allowing broader accessibility for researchers to conduct advanced brain studies.
Source: Allen Institute
Scientists have long known that our brains are organized into specialized areas, each responsible for distinct tasks. The visual cortex processes what we see, for instance, while the motor cortex governs movement. But how these regions form—and how their neural building blocks differ—remain a mystery.
A study published today in Nature sheds new light on the brain’s cellular landscape. Researchers at the Allen Institute for Brain Science used an advanced method called BARseq to swiftly classify and map millions of neurons across nine mouse brains.
They discovered that while brain regions share the same types of neurons, the specific combination of these cells gives each area a distinct ‘signature,’ akin to a cellular ID card.
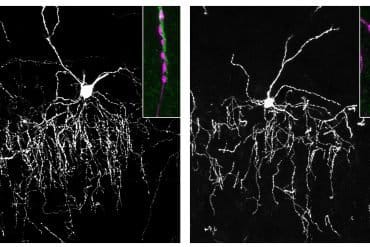
The team further explored how sensory inputs influence these cellular signatures. They discovered that mice deprived of sight experienced a major reorganization of cell types within the visual cortex, which blurred the distinctions with neighboring areas.
These shifts were not confined to the visual area but occurred across half of cortical regions, though to a lesser extent.
The study underscores the pivotal role of sensory experiences in shaping and maintaining each brain region’s unique cellular identity.
“BARseq lets us see with unprecedented precision how sensory inputs affect brain development,” said Xiaoyin Chen, Ph.D., the study’s co-lead author and an Assistant Investigator at the Allen Institute.
“These broad changes illustrate how important vision is in shaping our brains, even at the most basic level.”
A powerful new brain mapping tool
Previously, capturing single-cell data across multiple brains was challenging, said Mara Rue, Ph.D., co-lead author and a Scientist at the Allen Institute. But BARseq is cheaper and less time-consuming than similar mapping technologies, she said, enabling researchers to examine and compare brain-wide molecular architecture across multiple individuals.
BARseq tags individual brain cells with unique RNA ‘barcodes’ to track their connections across the brain. This data, combined with gene expression analysis, allows scientists to pinpoint and identify vast numbers of neurons in tissue slices.
For this study, the researchers used BARseq as a standalone method to rapidly analyze gene expression in intact tissue samples. In just three weeks, the researchers mapped more than 9 million cells from eight brains.
The scale and speed of BARseq provides scientists with a powerful new tool to delve deeper into the intricacies of the brain, Chen said.
“BARseq allows us to move beyond mapping what a ‘model’ or ‘standard’ brain looks like and start to use it as a tool to understand how brains change and vary,” Chen said. “With this throughput, we can now ask these questions in a very systematic way, something unthinkable with other techniques.”
Chen and Rue emphasized that the BARseq method is freely available. They hope their study encourages other researchers to use it to investigate the brain’s organizational principles or zoom in on cell types associated with disease.
“This isn’t something that only the big labs can do,” Rue said. “Our study is a proof of principle that BARseq allows a wide range of people in the field to use spatial transcriptomics to answer their own questions.”
About this brain mapping and neurotech research news
Author: Peter Kim
Source: Allen Institute
Contact: Peter Kim – Allen Institute
Image: The image is credited to Neuroscience News
Original Research: Open access.
“Whole-cortex in situ sequencing reveals input-dependent area identity” by Xiaoyin Chen et al. Nature
Abstract
Whole-cortex in situ sequencing reveals input-dependent area identity
The cerebral cortex is composed of neuronal types with diverse gene expression that are organized into specialized cortical areas. These areas, each with characteristic cytoarchitecture, connectivity and neuronal activity, are wired into modular networks.
However, it remains unclear whether these spatial organizations are reflected in neuronal transcriptomic signatures and how such signatures are established in development.
Here we used BARseq, a high-throughput in situ sequencing technique, to interrogate the expression of 104 cell-type marker genes in 10.3 million cells, including 4,194,658 cortical neurons over nine mouse forebrain hemispheres, at cellular resolution. De novo clustering of gene expression in single neurons revealed transcriptomic types consistent with previous single-cell RNA sequencing studies. The composition of transcriptomic types is highly predictive of cortical area identity.
Moreover, areas with similar compositions of transcriptomic types, which we defined as cortical modules, overlap with areas that are highly connected, suggesting that the same modular organization is reflected in both transcriptomic signatures and connectivity.
To explore how the transcriptomic profiles of cortical neurons depend on development, we assessed cell-type distributions after neonatal binocular enucleation.
Notably, binocular enucleation caused the shifting of the cell-type compositional profiles of visual areas towards neighbouring cortical areas within the same module, suggesting that peripheral inputs sharpen the distinct transcriptomic identities of areas within cortical modules.
Enabled by the high throughput, low cost and reproducibility of BARseq, our study provides a proof of principle for the use of large-scale in situ sequencing to both reveal brain-wide molecular architecture and understand its development.