Summary: A neurotoxin found in snake venom allowed researchers to uncover a detailed shape of nicotinic receptors involved in muscle contractions.
Source: UT Southwestern Medical Center
Researchers at UT Southwestern Medical Center have uncovered the detailed shape of a key protein involved in muscle contraction. The report, published today in Neuron, may lead to improved understanding of muscle-weakening genetic conditions called congenital myasthenic syndromes (CMS).
The protein sits on the surface of nerve cells that connect to muscles and is integral to triggering the muscle cell to contract just milliseconds after instructions are sent through the spinal cord. Called a nicotinic receptor, it has been a challenge to study because it sits in the cell’s membrane.
“The nicotinic receptor at the neuromuscular junction has been a target of interest for over a century. It was the first ion channel to be purified, the first to have its genes cloned, and the first to be imaged by an electron microscope,” says Ryan Hibbs, Ph.D., associate professor of Neuroscience and Biophysics at UTSW and a corresponding author of the study.
Many groups had tried to determine the receptor’s structure using an earlier technology called X-ray crystallography as well as first generation cryogenic electron microscopes (cryo-EM) but they were only able to obtain low-resolution images, he adds.
Normally the nicotinic receptor is activated by a molecule called acetylcholine. However, the nicotinic receptor is also the target of various venoms that cause muscle paralysis.
So the team used this to their advantage to isolate enough of the receptor protein to study its shape and structure. They mixed the toxin from snake venom with fish tissue known to contain high amounts of the receptor protein.
The team then flash froze the receptor bound to the toxin and used the rapidly evolving technique of cryo-electron microscopy to uncover the shape of the structure. Before recent developments in cryo-EM, the only method by which to solve protein structures like the nicotinic receptor was X-ray crystallography, which involved slowly growing crystals of proteins. But proteins that sit in membranes usually do not crystallize well.
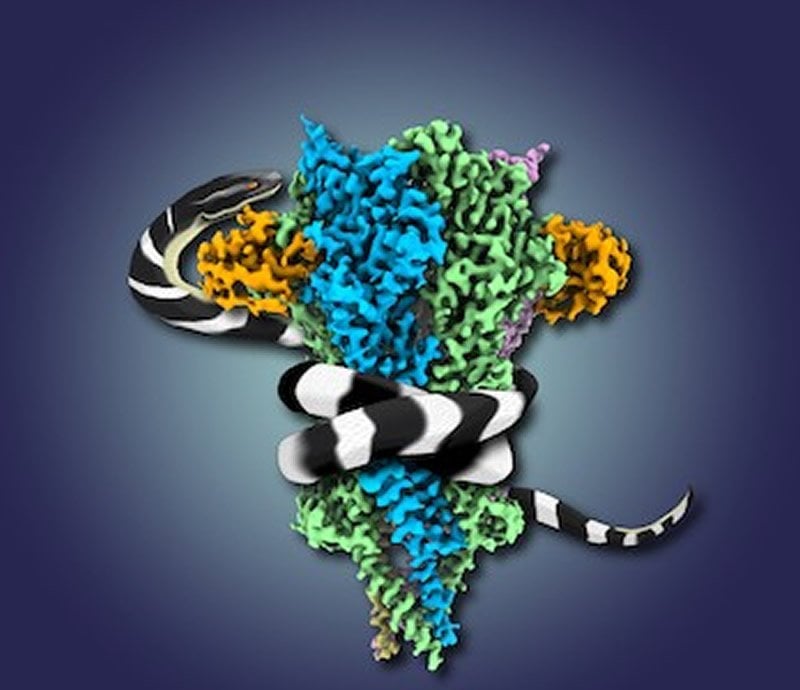
“This never could have been done using X-ray crystallography – hundreds of researchers had attempted it. The new microscopes allowed us to get to a very high resolution near the atomic level. At that resolution, we can precisely make out the positions of most of more than 2,000 amino acids that make up the receptor protein,” Hibbs says. “The results were stunning. They revealed, at a very fine level of detail, the 3D architecture of the receptor, with two toxin molecules bound to it exactly where we know the much smaller acetylcholine binds.”
The results showed how the toxin blocks acetylcholine by competing for the same binding site, paralyzing the receptor in the closed configuration and preventing the flow of electrochemical messages. The structure also reveals why the toxin binds so tightly and selectively to the receptor and how the venom paralyzes prey, including humans, who are unfortunate enough to get bitten by the snake.
“We learned a tremendous amount from the new 3D structure beyond how the toxin works to poison the receptor,” Hibbs says. Lead author Md. Mahfuzur Rahman, Ph.D., a postdoctoral researcher in the Hibbs lab, adds that the nicotinic acetylcholine neuromuscular receptor is a therapeutic target for several genetic diseases that cause muscle weakness, which the team also investigated.
“Here we mapped the mutations related to CMS to three principal regions on the receptor structure. This new structural information sheds light on how mutations in those areas result in muscle-weakening syndromes,” Rahman says.
UTSW co-authors include Jinfeng Teng and Colleen Noviello. Co-authors from the University of Colorado, Boulder, include co-corresponding author Michael H.B. Stowell, Brady Worrell, and Myeongseon Lee. Arthur Karlin of Columbia University also participated.
Funding: The research received support from the Cancer Prevention and Research Institute of Texas (grant No. RP170644), the National Institutes of Health (grant Nos. GM12957, DA037492, DA42072, NS095899, AG061829) and the MCDB Neurodegenerative Disease Fund. The authors declare no competing interests.
Hibbs is an Effie Marie Cain Scholar in Medical Research.
About this neuroscience research article
Source:
UT Southwestern Medical Center
Media Contacts:
Newsroom – UT Southwestern Medical Center
Image Source:
The image is credited to UT Southwestern.
Original Research: Open access
“Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins”. Md. Mahfuzur Rahman, Jinfeng Teng, Brady T. Worrell, Colleen M. Noviello, Myeongseon Lee, Arthur Karlin, Michael H.B. Stowell, Ryan E. Hibbs.
Neuron doi:10.1016/j.neuron.2020.03.012.
Abstract
Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins
Highlights
• High-resolution structure of a native muscle-type nicotinic acetylcholine receptor
• Previously unresolved structural elements contribute to neurotoxin binding
• Channel is stabilized in a closed conformation by α-bungarotoxin from snake venom
• Transduction mechanism and myasthenic disease mutations
Summary
The nicotinic acetylcholine receptor, a pentameric ligand-gated ion channel, converts the free energy of binding of the neurotransmitter acetylcholine into opening of its central pore. Here we present the first high-resolution structure of the receptor type found in muscle-endplate membrane and in the muscle-derived electric tissues of fish. The native receptor was purified from Torpedo electric tissue and functionally reconstituted in lipids optimal for cryo-electron microscopy. The receptor was stabilized in a closed state by the binding of α-bungarotoxin. The structure reveals the binding of a toxin molecule at each of two subunit interfaces in a manner that would block the binding of acetylcholine. It also reveals a closed gate in the ion-conducting pore, formed by hydrophobic amino acid side chains, located ∼60 Å from the toxin binding sites. The structure provides a framework for understanding gating in ligand-gated channels and how mutations in the acetylcholine receptor cause congenital myasthenic syndromes.
Feel Free To Share This Neuroscience News.