Summary: A new convolutional neural network that utilizes MRI brain scans can forecast genetic mutations in glioma brain tumors.
Source: Osaka University
Researchers at Osaka University have developed a computer method that uses magnetic resonance imaging (MRI) and machine learning to rapidly forecast genetic mutations in glioma tumors, which occur in the brain or spine. The work may help glioma patients to receive more suitable treatment faster, giving better outcomes. The research was recently published in Scientific Reports.
Cancer treatment has undergone a revolution in recent years. Spurred by recognition that each cancer case is unique, the specific genetic mutations tumor cells carry are now sequenced to discover which chemotherapy drugs will work best. However, certain types of cancer, especially brain tumors, are less accessible for genetic testing. The tumor’s genotype can’t be found until a sample is taken during surgery, and this can significantly delay treatment.
Glioma is a type of cancer that originates in the brain’s supporting cells. Two types of mutations are especially important; these are changes in the gene for the enzyme isocitrate dehydrogenase (IDH) or the promoter region of telomerase (TERT). Identifying these mutations can help direct the proper course of treatment. The researchers produced a machine-learning algorithm that can predict which mutations are present using only the MR images of the tumors.
“Machine learning is increasingly used to diagnose medical images. But our work is one of the first to even attempt to classify something as hidden as the genotype based on image data alone,” study first author Ryohei Fukuma explains. The algorithm was found to be significantly better at predicting the mutations compared with conventionally used radiomic features of the MR images, such as size, shape, and intensity.
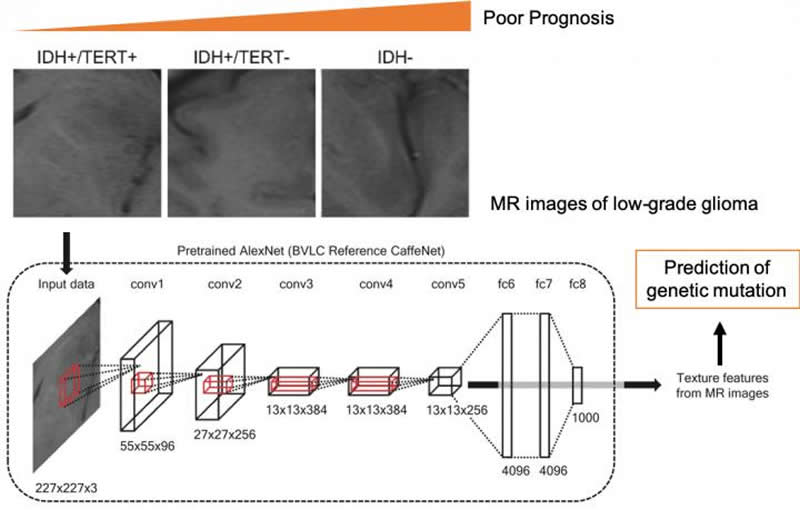
To construct the algorithm, the researchers used a convolutional neural network to extract features from the MR images. Then, using a machine-learning method called support vector machines, they classified the patients into groups based on the presence or absence of mutations. “We hope to expand this approach to other types of cancer, so we can take advantage of the large cancer gene databases already collected” senior author Haruhiko Kishima says.
The end result could remove the need for surgical tissue sampling. Even more, it could lead to better clinical outcomes for patients as the process of delivering personalized medicine becomes easier and faster.
Source:
Osaka University
Media Contacts:
Saori Obayashi – Osaka University
Image Source:
The image is credited to Osaka University.
Original Research: Open access
“Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network”. Ryohei Fukuma, Takufumi Yanagisawa, Manabu Kinoshita, Takashi Shinozaki, Hideyuki Arita, Atsushi Kawaguchi, Masamichi Takahashi, Yoshitaka Narita, Yuzo Terakawa, Naohiro Tsuyuguchi, Yoshiko Okita, Masahiro Nonaka, Shusuke Moriuchi, Masatoshi Takagaki, Yasunori Fujimoto, Junya Fukai, Shuichi Izumoto, Kenichi Ishibashi, Yoshikazu Nakajima, Tomoko Shofuda, Daisuke Kanematsu, Ema Yoshioka, Yoshinori Kodama, Masayuki Mano, Kanji Mori, Koichi Ichimura, Yonehiro Kanemura & Haruhiko Kishima.
Scientific Reports doi:10.1038/s41598-019-56767-3..
Abstract
Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network
Identification of genotypes is crucial for treatment of glioma. Here, we developed a method to predict tumor genotypes using a pretrained convolutional neural network (CNN) from magnetic resonance (MR) images and compared the accuracy to that of a diagnosis based on conventional radiomic features and patient age. Multisite preoperative MR images of 164 patients with grade II/III glioma were grouped by IDH and TERT promoter (pTERT) mutations as follows: (1) IDH wild type, (2) IDH and pTERT co-mutations, (3) IDH mutant and pTERT wild type. We applied a CNN (AlexNet) to four types of MR sequence and obtained the CNN texture features to classify the groups with a linear support vector machine. The classification was also performed using conventional radiomic features and/or patient age. Using all features, we succeeded in classifying patients with an accuracy of 63.1%, which was significantly higher than the accuracy obtained from using either the radiomic features or patient age alone. In particular, prediction of the pTERT mutation was significantly improved by the CNN texture features. In conclusion, the pretrained CNN texture features capture the information of IDH and TERT genotypes in grade II/III gliomas better than the conventional radiomic features.






