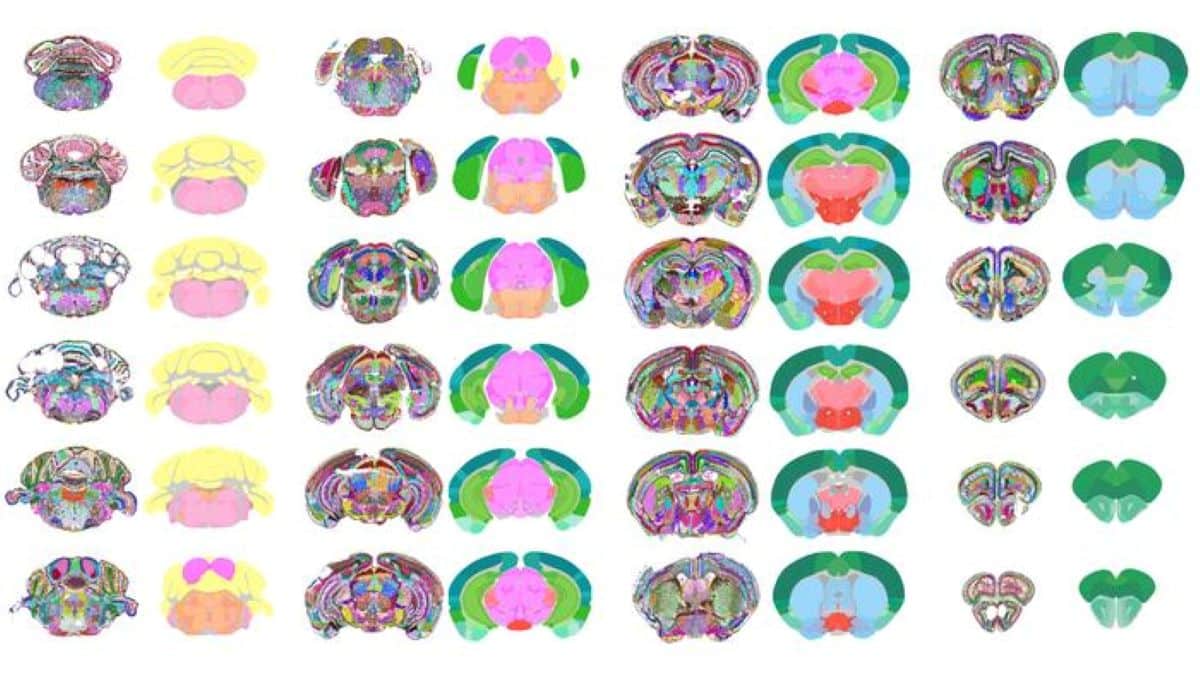
Summary: Researchers have created one of the most detailed maps of the mouse brain ever made, using artificial intelligence to reveal 1,300 distinct regions and subregions. The AI model, called CellTransformer, identified new brain areas that had never been charted before, providing an unprecedented view of brain organization.
Unlike traditional brain maps based on human interpretation, this one is entirely data-driven, drawing boundaries from molecular and cellular similarities. Beyond neuroscience, the model’s powerful “transformer” architecture could be applied to map other organs and even cancers, offering new insights into health and disease.
Key Facts:
- AI Brain Mapping: CellTransformer analyzed massive spatial transcriptomics data to reveal 1,300 mouse brain subregions.
- Data-Driven Discovery: The map is defined by real molecular and cellular data, not human interpretation.
- Broad Potential: The same AI framework could help map other organs, including cancer tissue, with similar precision.
Source: Allen Institute
In a powerful fusion of AI and neuroscience, researchers at the University of California, San Francisco (UCSF) and Allen Institute designed an AI model that has created one of the most detailed maps of the mouse brain to date, featuring 1,300 regions/subregions.
This new map includes previously uncharted subregions of the brain, opening new avenues for neuroscience exploration.
The findings were published today in Nature Communications.
They offer an unprecedented level of detail and advance our understanding of the brain by allowing researchers to link specific functions, behaviors, and disease states to smaller, more precise cellular regions—providing a roadmap for new hypotheses and experiments about the roles these areas play.
“It’s like going from a map showing only continents and countries to one showing states and cities,” said Bosiljka Tasic, Ph.D., director of molecular genetics at the Allen Institute and one of the study authors.
“This new, detailed brain parcellation solely based on data, and not human expert annotation, reveals previously uncharted subregions of the mouse brain. And based on decades of neuroscience, new regions correspond to specialized brain functions to be discovered.”
At the heart of this breakthrough is CellTransformer, a powerful AI model that can automatically identify important subregions of the brain from massive spatial transcriptomics datasets. Spatial transcriptomics reveals where certain brain cell types are positioned in the brain but does not reveal regions of the brain based on their composition.
Now, CellTransformer allows scientists to define brain regions and subdivisions based on calculations of shared cellular neighborhoods, much like sketching a city’s borders based on the types of buildings within it.
“Our model is built on the same powerful technology as AI tools like ChatGPT. Both are built on a ‘transformer’ framework which excels at understanding context,” said Reza Abbasi-Asl, Ph.D., associate professor of neurology and bioengineering at UCSF and senior author of the study.
“While transformers are often applied to analyze the relationship between words in a sentence, we use CellTransformer to analyze the relationship between cells that are nearby in space. It learns to predict a cell’s molecular features based on its local neighborhood, allowing it to build up a detailed map of the overall tissue organization.”
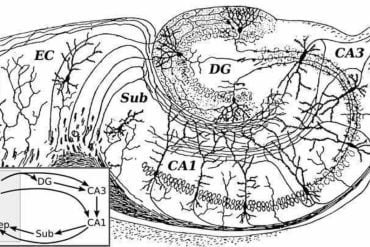
This model successfully replicates known regions of the brain, such as the hippocampus; but more importantly, it can also discover previously uncatalogued, finer-grained subregions in poorly understood brain regions, such as the midbrain reticular nucleus, which plays a complex role in movement initiation and release.
What Makes this Brain Map Distinct from Others
This new brain map depicts brain regions, versus cell types; and unlike previous brain maps, CellTransformer’s is entirely data-driven, meaning its boundaries are defined by cellular and molecular data rather than human interpretation. With 1,300 regions and subregions, it also represents one of the most granular and complex data-driven brain maps of any animal to date.
Role of the Allen Institute’s Common Coordinates Framework (CCF)
The Allen Institute’s Common Coordinate Framework (CCF) served as the essential gold standard for validating CellTransformer’s accuracy.
“By comparing the brain regions automatically identified by CellTransformer to the CCF, we were able to show that our data-driven method was identifying areas aligned with known expert-defined anatomical structures,” said Alex Lee, a PhD candidate at UCSF and first author of the study.
“Seeing that our model produces results so similar to CCF, which is such a well-characterized and high-quality resource for the field, was reassuring. The high level of agreement with the CCF provided a critical benchmark, giving confidence that the new subregions discovered by CellTransformer may also be biologically meaningful. We are hoping to explore and validate the results with further computational and experimental studies.”
The potential of this research to unlock critical insights reaches beyond neuroscience. CellTransformer’s powerful AI capabilities are tissue agnostic: They can be used on other organ systems and tissues, including cancerous tissue, where large-scale spatial transcriptomics data is available to better understand the biology of health and disease and fuel the discovery of new treatments and therapies.
Key Questions Answered:
A: Unlike previous brain maps, it defines brain regions purely from cellular and molecular data—without relying on human annotation—resulting in unprecedented accuracy and detail.
A: Built on the same transformer framework that powers language models, it analyzes spatial relationships between cells to infer how brain tissue is organized.
A: It provides a precise blueprint for studying brain function, disease, and cellular interactions—and could also be adapted to map other organs or tumor structures.
About this brain mapping and artificial intelligence research news
Author: Peter Kim
Source: Allen Institute
Contact: Peter Kim – Allen Institute
Image: The image is credited to UCSF
Original Research: Open access.
“Data-driven fine-grained region discovery in the mouse brain with transformers” by Bosiljka Tasic, et al. Nature Communications
Abstract
Data-driven fine-grained region discovery in the mouse brain with transformers
Spatial transcriptomics offers unique opportunities to define the spatial organization of tissues and organs, such as the mouse brain.
We address a key bottleneck in the analysis of organ-scale spatial transcriptomic data by establishing a workflow for self-supervised spatial domain detection that is scalable to multimillion-cell datasets.
This workflow uses a self-supervised framework for learning latent representations of tissue spatial domains or niches.
We use an encoder-decoder architecture, which we named CellTransformer, to hierarchically learn higher-order tissue features from lower-level cellular and molecular statistical patterns.
Coupling our representation learning workflow with minibatched GPU-accelerated clustering algorithms allows us to scale to multi-million cell MERFISH datasets where other methods cannot.
CellTransformer is effective at integrating cells across tissue sections, identifying domains highly similar to ones in existing ontologies such as Allen Mouse Brain Common Coordinate Framework (CCF) while allowing discovery of hundreds of uncataloged areas with minimal loss of domain spatial coherence.
CellTransformer domains recapitulate previous neuroanatomical studies of areas in the subiculum and superior colliculus and characterize putatively uncataloged subregions in subcortical areas, which currently lack subregion annotation.
CellTransformer is also capable of domain discovery in whole-brain Slide-seqV2 datasets. Our workflows enable complex multi-animal analyses, achieving nearly perfect consistency of up to 100 spatial domains in a dataset of four individual mice with nine million cells across more than 200 tissue sections.
CellTransformer advances the state of the art for spatial transcriptomics by providing a performant solution for the detection of fine-grained tissue domains from spatial transcriptomics data.