Summary: FlyEM, a team of scientists from Google and the Janelia Research Campus at Howard Hughes Medical Institute, has released the most complete map of the fly brain ever created. The map pinpoints millions of connections between 25,000 neurons. The researchers have made the data free online, along with all of the tools necessary to use it.
Source: Howard Hughes Medical Institute
In a darkened room in Ashburn, Virginia, rows of scientists sit at computer screens displaying vivid 3-D shapes. With a click of a mouse, they spin each shape to examine it from all sides. The scientists are working inside a concrete building at the Howard Hughes Medical Institute’s Janelia Research Campus, just off a street called Helix Drive. But their minds are somewhere else entirely – inside the brain of a fly.
Each shape on the scientists’ screens represents part of a fruit fly neuron. These researchers and others at Janelia are tackling a goal that once seemed out of reach: outlining each of the fly brain’s roughly 100,000 neurons and pinpointing the millions of places they connect. Such a wiring diagram, or connectome, reveals the complete circuitry of different brain areas and how they’re linked. The work could help unlock networks involved in memory formation, for example, or neural pathways that underlie movements.
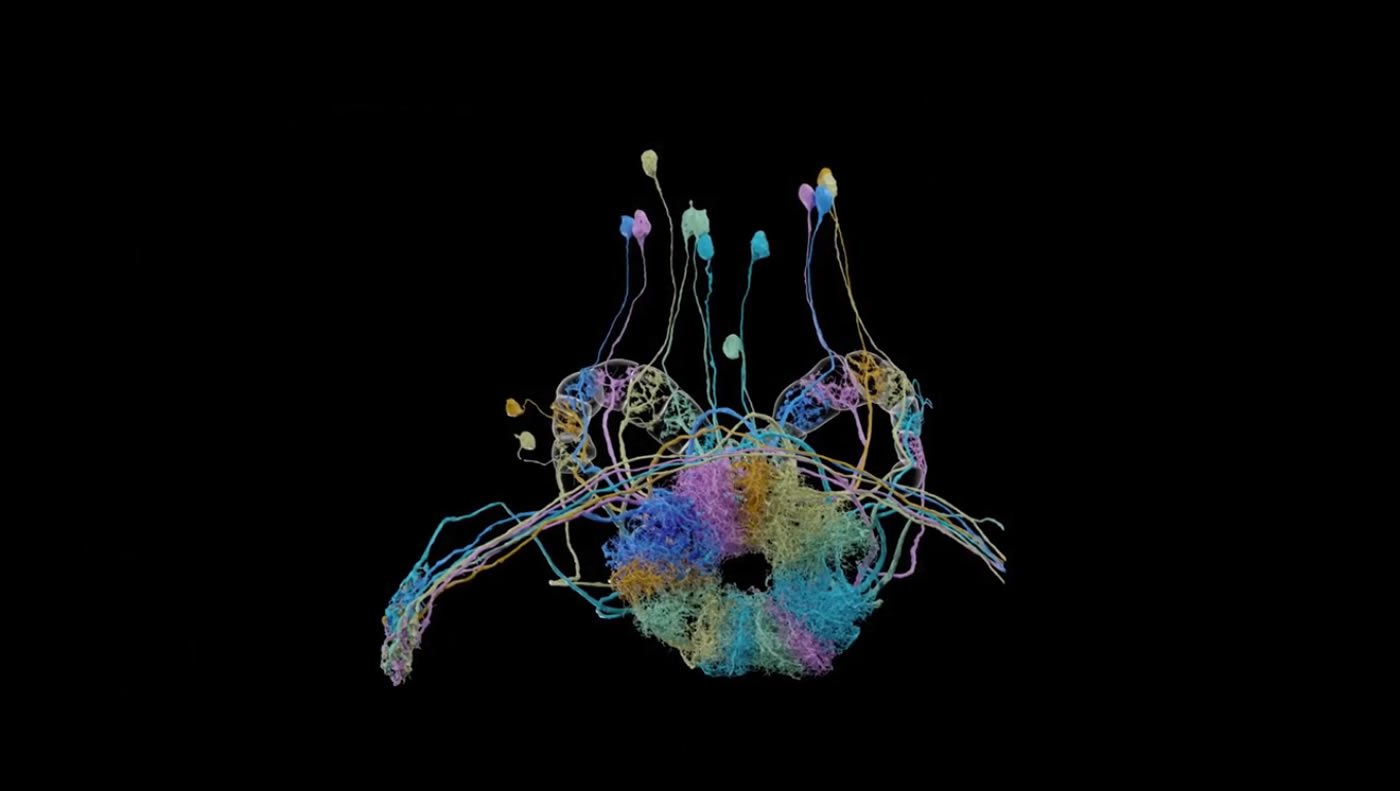
Gerry Rubin, vice president of HHMI and executive director of Janelia, has championed this project for more than a decade. It’s a necessary step in understanding how the brain works, he says. When the project began, Rubin estimated that with available methods, tracing the connections between every fly neuron by hand would take 250 people working for two decades – what he refers to as “a 5,000 person-year problem.”
Now, a stream of advances in imaging technology and deep-learning algorithms have yanked the dream of a fly connectome out of the clouds and into the realm of probability. High-powered customized microscopes, a team of dedicated neural proofreaders and data analysts, and a partnership with Google have sped up the process by orders of magnitude.
Completing the hemibrain connectome required optimizing every part of the process, from the microscopes that imaged the brain to the algorithms that automatically traced neurons through the dataset. This video gives an overview of the endeavor. Credit: HHMI.
Today, a team of Janelia researchers reports hitting a critical milestone: they’ve traced the path of every neuron in a portion of the female fruit fly brain they’ve dubbed the “hemibrain.” The map encompasses 25,000 neurons – roughly a third of the fly brain, by volume – but its impact is outsized. It includes regions of keen interest to scientists — those that control functions like learning, memory, smell, and navigation. With more than 20 million neural connections pinpointed so far, it’s the biggest and most detailed map of the fly brain ever completed.
The connectome project team, known as FlyEM, is making the data — and all of the tools necessary to use it — available for free. They describe the work in a paper posted to bioRxiv on January 21, 2020. And they’re currently on track to complete a connectome of the entire fly nervous system by 2022.
Source:
Howard Hughes Medical Institute
Media Contacts:
Meghan Rosen – Howard Hughes Medical Institute
Image Source:
The image is credited to FlyEM/Janelia Research Campus.
Original Research: Closed access
“A Connectome of the Adult Drosophila Central Brain”. C. Shan Xu, Michal Januszewski, Zhiyuan Lu, Shin-ya Takemura, Kenneth Hayworth, Gary Huang, Kazunori Shinomiya, Jeremy Maitin-Shepard, David Ackerman, Stuart Berg, Tim Blakely, John Bogovic, Jody Clements, Tom Dolafi, Philip Hubbard, Dagmar Kainmueller, William Katz, Takashi Kawase, Khaled Khairy, Laramie Leavitt, Peter H. Li, Larry Lindsey, Nicole Neubarth, Donald J. Olbris, Hideo Otsuna, Eric T. Troutman, Lowell Umayam, Ting Zhao, Masayoshi Ito, Jens Goldammer, Tanya Wolff, Robert Svirskas, Philipp Schlegel, Erika R. Neace, Christopher J. Knecht Jr., Chelsea X. Alvarado, Dennis Bailey, Samantha Ballinger, Jolanta A. Borycz, Brandon Canino, Natasha Cheatham, Michael Cook, Marisa Dreyer, Octave Duclos, Bryon Eubanks, Kelli Fairbanks, Samantha Finley, Nora Forknall, Audrey Francis, Gary Patrick Hopkins, Emily M. Joyce, SungJin Kim, Nicole A. Kirk, Julie Kovalyak, Shirley A. Lauchie, Alanna Lohff, Charli Maldonado, Emily A. Manley, Sari McLin, Caroline Mooney, Miatta Ndama, Omotara Ogundeyi, Nneoma Okeoma, Christopher Ordish, Nicholas Padilla, Christopher Patrick, Tyler Paterson, Elliott E. Phillips, Emily M. Phillips, Neha Rampally, Caitlin Ribeiro, Madelaine K. Robertson, Jon Thomson Rymer, Sean M. Ryan, Megan Sammons, Anne K. Scott, Ashley L. Scott, Aya Shinomiya, Claire Smith, Kelsey Smith, Natalie L. Smith, Margaret A. Sobeski, Alia Suleiman, Jackie Swift, Satoko Takemura, Iris Talebi, Dorota Tarnogorska, Emily Tenshaw, Temour Tokhi, John J. Walsh, Tansy Yang, Jane Anne Horne, Feng Li, Ruchi Parekh, Patricia K. Rivlin, Vivek Jayaraman, Kei Ito, Stephan Saalfeld, Reed George, Ian Meinertzhagen, Gerald M. Rubin, Harald F. Hess, Louis K. Scheffer, Viren Jain, Stephen M. Plaza.
bioRxiv doi:10.1101/2020.01.21.911859.
Abstract
A Connectome of the Adult Drosophila Central Brain
The neural circuits responsible for behavior remain largely unknown. Previous efforts have reconstructed the complete circuits of small animals, with hundreds of neurons, and selected circuits for larger animals. Here we (the FlyEM project at Janelia and collaborators at Google) summarize new methods and present the complete circuitry of a large fraction of the brain of a much more complex animal, the fruit fly Drosophila melanogaster. Improved methods include new procedures to prepare, image, align, segment, find synapses, and proofread such large data sets; new methods that define cell types based on connectivity in addition to morphology; and new methods to simplify access to a large and evolving data set. From the resulting data we derive a better definition of computational compartments and their connections; an exhaustive atlas of cell examples and types, many of them novel; detailed circuits for most of the central brain; and exploration of the statistics and structure of different brain compartments, and the brain as a whole. We make the data public, with a web site and resources specifically designed to make it easy to explore, for all levels of expertise from the expert to the merely curious. The public availability of these data, and the simplified means to access it, dramatically reduces the effort needed to answer typical circuit questions, such as the identity of upstream and downstream neural partners, the circuitry of brain regions, and to link the neurons defined by our analysis with genetic reagents that can be used to study their functions. Note: In the next few weeks, we will release a series of papers with more involved discussions. One paper will detail the hemibrain reconstruction with more extensive analysis and interpretation made possible by this dense connectome. Another paper will explore the central complex, a brain region involved in navigation, motor control, and sleep. A final paper will present insights from the mushroom body, a center of multimodal associative learning in the fly brain.






