Summary: Researchers have uncovered a mechanism that may trigger ALS’s earliest stages, identifying proteins that mislocalize, causing neuron degeneration. By targeting the RNA-binding protein SmD1, scientists were able to prevent key ALS proteins from leaving their protective cellular zones, preserving neuron function.
The findings may lead to ALS therapies capable of halting progression before significant neurodegeneration occurs, offering potential new strategies against the disease.
Key Facts:
- ALS progression may begin when the protein CHMP7 mislocalizes, initiating neuron damage.
- Restoring SmD1 function could prevent CHMP7 mislocalization and protect neurons.
- The team hopes that therapies like risdiplam may slow ALS onset by stabilizing neuron integrity.
Source: UCSD
Approximately 5,000 people in the U.S. develop amyotrophic lateral sclerosis (ALS) each year. On average, they survive for only two to five years after being diagnosed, according to the Centers for Disease Control and Prevention.
The rapidly progressing neurodegenerative disease causes the death of neurons in the brain and spinal cord, resulting in muscle weakness, respiratory failure and dementia.
Despite the devastating nature of the disease, little is known about what first triggers the deterioration of motor neurons at the onset of ALS.
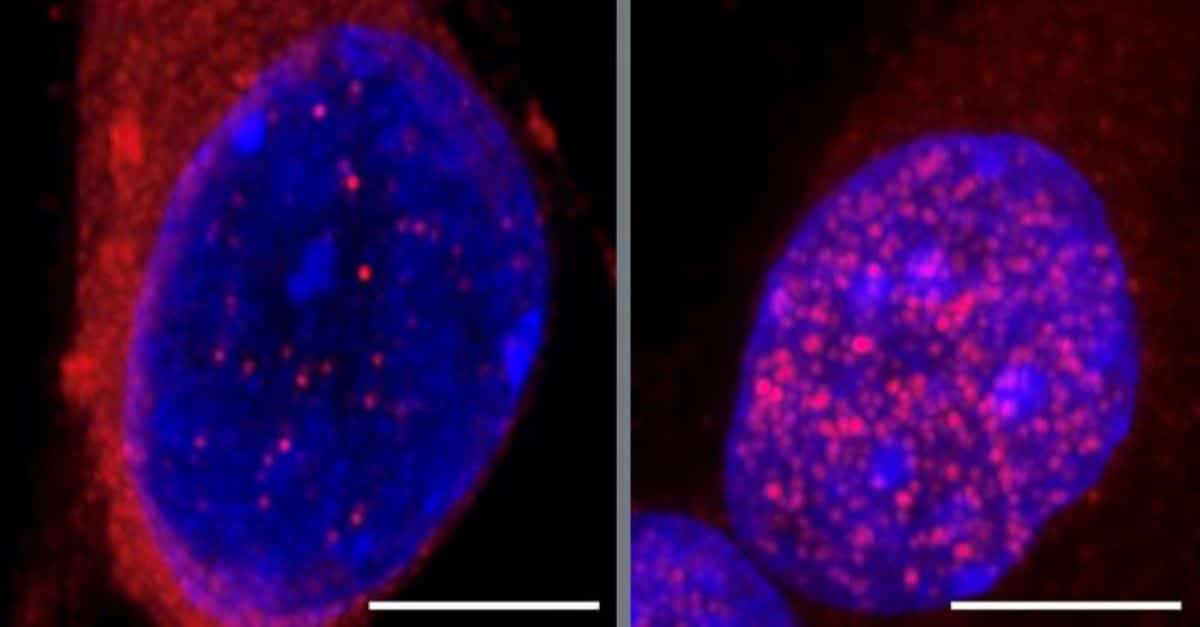
Now, researchers from University of California San Diego and their colleagues report that they have identified a key pathway that sets off neurodegeneration in the early stages of the disease.
The findings could lead to development of therapies to prevent or slow the progression of ALS early on, before major damage has been done.
The study was published on October 31, 2024 in Neuron.
A protein called TDP-43 is usually located in the nucleus of motor neurons, where it regulates gene expression required for the cells to function. Studies have shown that when TDP-43 instead accumulates in the cytoplasm, outside of the nucleus, it is a telltale sign of ALS. How the protein ends up in the wrong place, leading to neuronal degeneration, has stumped researchers until now.
“By the time you see a patient with ALS and you see the TDP-43 protein aggregated in the cytoplasm, it’s like the accident site with all the cars crashed already, but that’s not the initiating event,” said corresponding author Gene Yeo, Ph.D., professor in the Department of Cellular and Molecular Medicine at UC San Diego School of Medicine and director of the Center for RNA Technologies and Therapeutics and the Sanford Stem Cell Institute Innovation Center.
Tracing the events leading up to the “accident”, Yeo explains that another protein, called CHMP7 — normally found in the cytoplasm — builds up in the nucleus instead, setting off a cascade of events that ultimately lead to motor neuron degeneration. But what causes CHMP7 to accumulate in the nucleus to begin with?
Yeo and his team screened for RNA-binding proteins that might influence CHMP7 build-up in the nucleus. This yielded 55 proteins, 23 of which had a potential connection to ALS pathogenesis. Inhibiting the production of several of these proteins led to an increase in CHMP7 in the nucleus.
Further experiments with motor neurons created from ALS-patient-derived induced pluripotent stem cells resulted in the surprising discovery that depleting one of these, an RNA-splicing associated protein called SmD1, which had not previously been known to affect CHMP7 levels, led to the largest uptick in its nuclear accumulation.
A build-up of CHMP7 in the nucleus damages nucleoporins, which Yeo likens to tiny portals in the membrane separating the nucleus from the cytoplasm that orchestrate the movement of proteins and RNA between the two cellular spaces. Dysfunctional nucleoporins allow TDP-43 to exit the nucleus and accumulate in the cytoplasm. Once there, the protein can no longer oversee the gene expression programs necessary for neurons to function.
However, when the researchers boosted SmD1 expression in cells, CHMP7 was restored to its usual location in the cytoplasm, leaving nucleopores intact, allowing TDP-43 to stay in the nucleus, thus sparing the motor neurons from degeneration.
“You can actually fix the localization of this CHMP7 protein and therefore all of the downstream effects,” said Norah Al-Azzam, first author of the study, then a neurosciences student in the Yeo lab who went on to earn her Ph.D. in spring 2024.
What’s more, the SmD1 protein is part of SMN, a multiprotein complex. SMN dysfunction is implicated in another neurodegenerative disorder, spinal muscular atrophy.
“We’re intrigued because there are actually therapeutics for spinal muscular atrophy,” Yeo said. “One of them, risdiplam, is a small molecule compound that enhances the splicing and expression of SMN2, a gene closely related to the SMN1 gene that becomes dysfunctional in ALS.”
This hints at the possibility that using risdiplam to raise SMN levels could prevent ALS from developing past the earliest stage of the disease.
“It’s not like all the neurons die at once,” said Yeo. “Some neurons die first, and then there is spread across other neurons. Perhaps as soon as you get symptoms, we could treat the patient so that the rest of the neurons don’t crash and hope you stop the progression of ALS.”
The researchers think the SMN complex could play a crucial role in the onset of ALS, but further research is needed. The next steps will be to raise funds to continue the research in animal models and in other genetic models of ALS, and eventually test the effectiveness of risdiplam or other compounds for short-circuiting ALS.
Additional co-authors on the study include: Jenny H. To, Vaishali Gautam, Dylan C. Lam, Chloe B. Nguyen, Jack T. Naritomi, Assael A. Madrigal, Benjamin Lee, Anthony Avina, Orel Mizrahi, Jasmine R. Mueller, Willard Ford, Anthony Q. Vu, Steven M. Blue, Yashwin L. Madakamutil, Uri Manor, Cara R. Schiavon and Elena Rebollo, all at UC San Diego; Wenhao Jin at Sanford Laboratories for Innovative Medicines; Lena A. Street and Marko Jovanovic at Columbia University; Jeffrey D. Rothstein and Alyssa N. Coyne at Johns Hopkins University School of Medicine.
Funding: The study was funded, in part, by the National Institutes of Health (grants R01HG004659, U24 HG009889, R35GM128802, R01AG071869 and R01HG012216), the National Science Foundation (MCB-2224211), and the Chan-Zuckerberg Initiative.
About this ALS research news
Author: Susanne Bard
Source: UCSD
Contact: Susanne Bard – UCSD
Image: The image is credited to UCSD
Original Research: Open access.
“Inhibition of RNA splicing triggers CHMP7 nuclear entry, impacting TDP-43 function and leading to the onset of ALS cellular phenotypes” by Gene Yeo et al. Neuron
Abstract
Inhibition of RNA splicing triggers CHMP7 nuclear entry, impacting TDP-43 function and leading to the onset of ALS cellular phenotypes
Amyotrophic lateral sclerosis (ALS) is linked to the reduction of certain nucleoporins in neurons. Increased nuclear localization of charged multivesicular body protein 7 (CHMP7), a protein involved in nuclear pore surveillance, has been identified as a key factor damaging nuclear pores and disrupting transport.
Using CRISPR-based microRaft, followed by gRNA identification (CRaft-ID), we discovered 55 RNA-binding proteins (RBPs) that influence CHMP7 localization, including SmD1, a survival of motor neuron (SMN) complex component.
Immunoprecipitation-mass spectrometry (IP-MS) and enhanced crosslinking and immunoprecipitation (CLIP) analyses revealed CHMP7’s interactions with SmD1, small nuclear RNAs, and splicing factor mRNAs in motor neurons (MNs).
ALS induced pluripotent stem cell (iPSC)-MNs show reduced SmD1 expression, and inhibiting SmD1/SMN complex increased CHMP7 nuclear localization.
Crucially, overexpressing SmD1 in ALS iPSC-MNs restored CHMP7’s cytoplasmic localization and corrected STMN2 splicing. Our findings suggest that early ALS pathogenesis is driven by SMN complex dysregulation.






