Summary: Researchers created the largest 3D reconstruction of human brain tissue at synaptic resolution, capturing detailed images of a cubic millimeter of human temporal cortex. This tiny piece of brain contains 57,000 cells, 230 millimeters of blood vessels, and 150 million synapses, which amounts to 1,400 terabytes of data.
This research is part of a broader effort to map an entire mouse brain’s neural wiring, with hopes of advancing our understanding of brain function and disease. The technology combines high-resolution electron microscopy and AI-powered algorithms to meticulously color-code and map out the complex neural connections.
Key Facts:
- Intricate Details: The reconstructed brain tissue includes unprecedented details, such as a unique set of axons connected by up to 50 synapses, and other peculiar formations potentially linked to epilepsy in the patient from whom the sample was taken.
- Massive Data Volume: The study tackled a piece of brain tissue containing a data volume equivalent to 1,400 terabytes, illustrating the complexity and data-intensive nature of mapping neural connections.
- Technological Integration: The research utilized cutting-edge AI algorithms developed by Google to assist in the 3D reconstruction of the brain tissue, highlighting a significant collaboration between biological research and technological innovation.
Source: Harvard
A cubic millimeter of brain tissue may not sound like much. But considering that tiny square contains 57,000 cells, 230 millimeters of blood vessels, and 150 million synapses, all amounting to 1,400 terabytes of data, Harvard and Google researchers have just accomplished something enormous.
A Harvard team led by Jeff Lichtman, the Jeremy R. Knowles Professor of Molecular and Cellular Biology and newly appointed dean of science, has co-created with Google researchers the largest synaptic-resolution, 3D reconstruction of a piece of human brain to date, showing in vivid detail each cell and its web of neural connections in a piece of human temporal cortex about half the size of a rice grain.
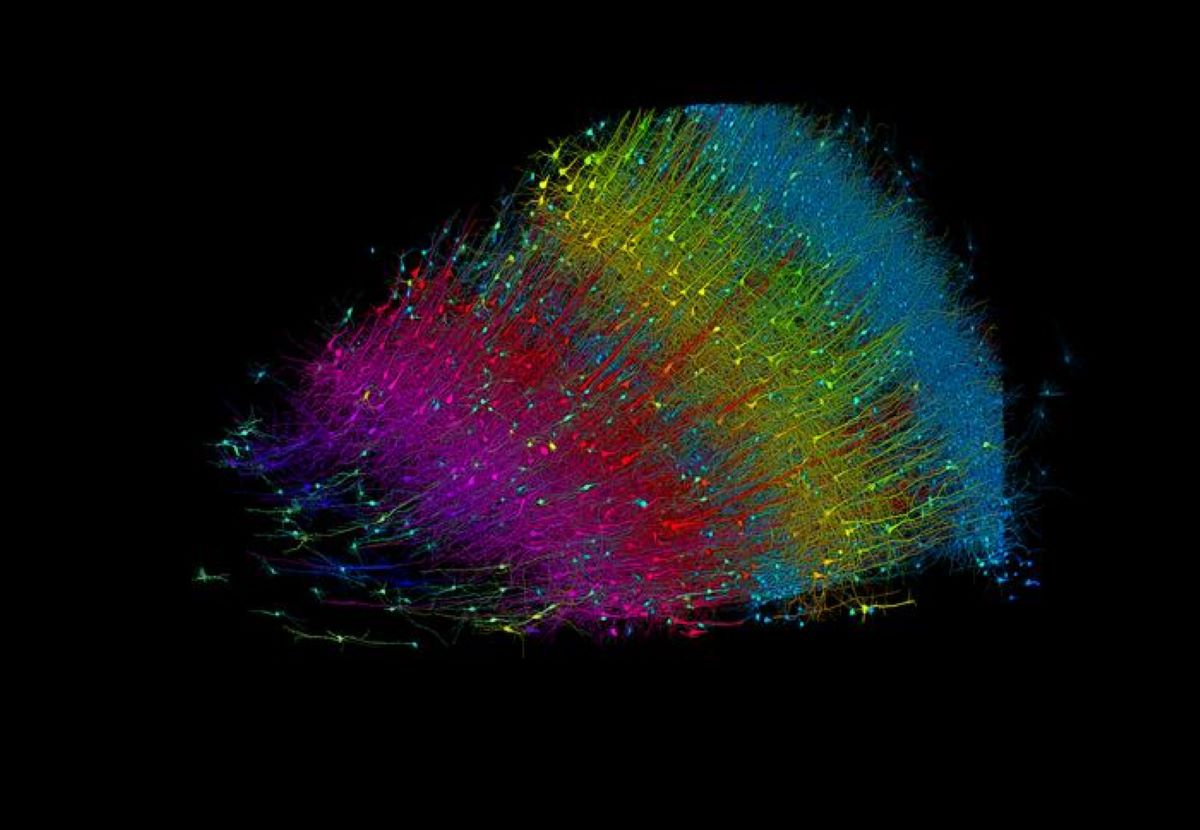
The feat, published in Science, is the latest in a nearly 10-year collaboration with scientists at Google Research, who combine Lichtman’s electron microscopy imaging with AI algorithms to color-code and reconstruct the extremely complex wiring of mammal brains.
The paper’s three co-first authors are former Harvard postdoctoral researcher Alexander Shapson-Coe; Michał Januszewski of Google Research, and Harvard postdoctoral researcher Daniel Berger.
The collaboration’s ultimate goal, supported by the National Institutes of Health BRAIN Initiative, is to create a high-resolution map of a whole mouse brain’s neural wiring, which would entail about 1,000 times the amount of data they just produced from the 1-cubic-millimeter fragment of human cortex.
“The word ‘fragment’ is ironic,” Lichtman said. “A terabyte is, for most people, gigantic, yet a fragment of a human brain – just a miniscule, teeny-weeny little bit of human brain – is still thousands of terabytes.”
The latest map in Science contains never-before-seen details of brain structure, including a rare but powerful set of axons connected by up to 50 synapses. The team also noted oddities in the tissue, such as a small number of axons that formed extensive whorls. Since their sample was taken from a patient with epilepsy, they’re unsure if such unusual formations are pathological or simply rare.
Lichtman’s field is “connectomics,” which, analogous to genomics, seeks to create comprehensive catalogues of brain structure, down to individual cells and wiring. Such completed maps would light the way toward new insights into brain function and disease, about which scientists still know very little.
Google’s state-of-the-art AI algorithms allow for reconstruction and mapping of brain tissue in three dimensions. The team has also developed a suite of publicly available tools researchers can use to examine and annotate the connectome.
“Given the enormous investment put into this project, it was important to present the results in a way that anybody else can now go and benefit from them,” said Google Research collaborator Viren Jain.
Next the team will tackle the mouse hippocampal formation, which is important to neuroscience for its role in memory and neurological disease.
About this brain mapping research news
Author: Anne Manning
Source: Harvard
Contact: Anne Manning – Harvard
Image: The image is credited to Google Research and Lichtman Lab
Original Research: Closed access.
“A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution” by Jeff Lichtman et al. Science
Abstract
A petavoxel fragment of human cerebral cortex reconstructed at nanoscale resolution
INTRODUCTION
Although the functions performed by most of the vital organs in humans are not very different compared with other animals, those performed by the human brain clearly separate us from the rest of life on the planet. However, detailed knowledge concerning the synaptic circuitry underlying human brain function is lacking.
Connectomic imaging approaches are now available to render neural circuits of sufficiently large volume and high enough resolution to study the connectivity at the level of individual neurons and their synaptic connections but over a scale comprising thousands of neurons. Generating such a dataset was the goal of this project.
RATIONALE
One critical barrier to obtaining human neural circuits has been the access to high-quality human brain tissue. Organ biopsies provide valuable information in many human organ systems, but biopsies are rarely done in the brain except to examine or excise neoplastic masses, so most of them are problematic for the investigation of normal human brain structure.
One attempt has been to use brain organoids made from human cells, but at present, they do not approximate brain tissue architectonics (e.g., cortical layers are not present).
A direct approach would be to map cells and circuits from human specimens made available from neurosurgical interventions for neurological conditions in which pieces of the cortex are discarded because they obstruct access to a pathological site.
We posited that the human brain tissue that is a by-product of neurosurgical procedures could be leveraged to study normal—and ultimately disordered—human neural circuits.
RESULTS
Here, we describe such a sample of human temporal cortex, 1 mm3 in volume, which extends through all cortical layers. The sample was obtained during surgery to gain access to an underlying hippocampal lesion from a patient with epilepsy.
We imaged this sample by high-throughput serial section electron microscopy, generating a petascale dataset that was analyzed with new tools and computationally intensive methods.
We reconstructed thousands of neurons, more than a hundred million synaptic connections, and all of the other tissue elements that comprise human brain matter, including glial cells, the blood vasculature, and myelin.
Because the dataset is large and incompletely scrutinized, we are sharing all of the data in an online resource (https://h01-release.storage.googleapis.com/landing.html) and also providing tools for analysis and proofreading.
We found a previously unrecognized class of directionally oriented neurons in deep layers (see figure, panel J) and very powerful and rare multisynaptic connections between neurons throughout the sample (see figure, panel K).
CONCLUSION
This work provides evidence of the feasibility of human connectomic approaches to visualize and ultimately gain insight into the physical underpinnings of normal and disordered human brain function. It is hoped that this endeavor will be aided by providing free access to all of the data and relevant tools.







