Summary: A new imaging technique captures information about the function and structure of brain tissue at the subcellular level.
Source: Francis Crick Institute
Researchers at the Francis Crick Institute have developed an imaging technique to capture information about the structure and function of brain tissue at subcellular level – a few billionth of a metre, while also capturing information about the surrounding environment.
The unique approach detailed in Nature Communications today overcomes the challenges of imaging tissues at different scales, allowing scientists to see the surrounding cells and how they function, so they can build a complete picture of neural networks in the brain.
Various imaging methods are used to capture information about tissue, cells and subcellular structures. However, a single method can only capture information about either the structure or function of the tissue and looking in detail at a nanometre scale means scientists lose information about the wider surroundings. This means that to gain an overall understanding of the tissue, imaging techniques need to be combined.
In their study, the scientists developed an approach which combines seven imaging methods, including in vivo imaging, synchrotron X-ray, and volume electron microscopy. They demonstrated their approach by imaging two different areas of the brain in mice – the olfactory bulb and the hippocampus.
Importantly, the technique could be applied to other areas of the brain or parts of the body, providing scientists with a more detailed understanding of many different biological structures and tissues.
Each step of the imaging process provides different information. Firstly, the researchers used in vivo calcium imaging to visualise neurons in specific regions of the brain and see which neurons were active when the mice were exposed to odours.
After the mice were euthanised, brain tissue samples were imaged using various methods including synchrotron X-ray tomography, which captures samples up to several millimetres in length.
This scale is sufficient for scientists to see whole neural networks and also where particular cells or other structures sit within the wider context of the sample. Importantly, this method does not damage the sample so it can be imaged again using another technique.
The researchers then selected areas of particular interest to be imaged with electron microscopy, capturing intricate detail at a high resolution. At some target areas this could map details as small as 10 nanometres, allowing researchers to see tiny structures like the individual synapses that connect neurons.
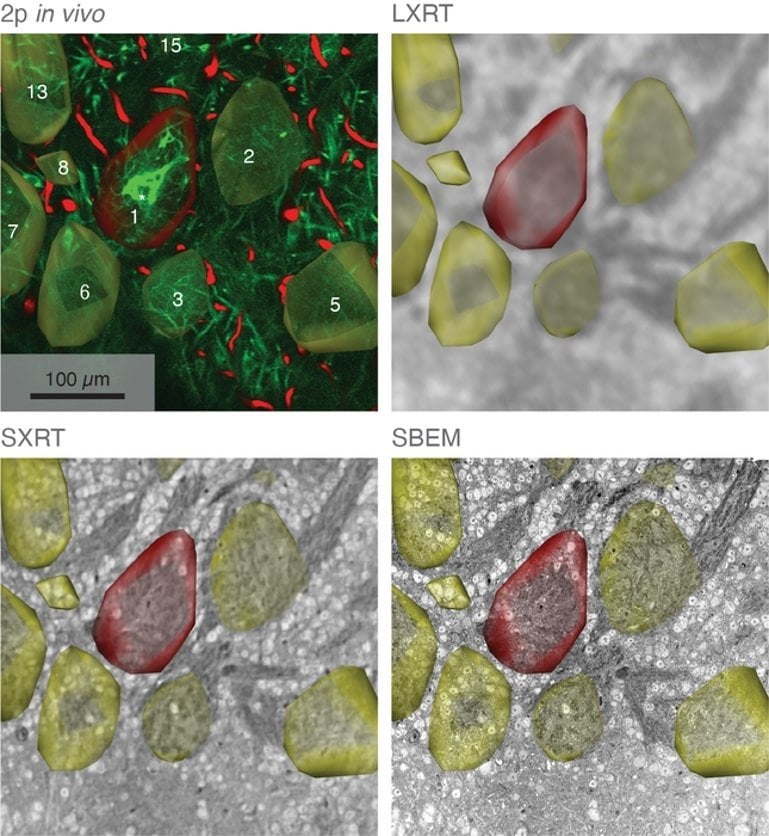
Using computer algorithms, they combined the results to create a complete map of the structure and function of the section of the brain they were studying, up to a few cubic millimetres.
Carles Bosch, first author and Principal Laboratory Research Scientist in the Sensory Circuits and Neurotechnology Laboratory at the Crick says: “Our approach provides a reliable way to overcome the challenge of imaging structures at vastly different scales. We believe it will be a powerful tool to investigate neuronal circuits in the brains of mammals as well as the structure and function of other tissues.”
Andreas Schaefer, senior author and head of the Sensory Circuits and Neurotechnology Laboratory at the Crick says: “We’re interested in applying this approach to the brain, where it is important to gather information about whole neural networks which are several millimetres in length alongside information about specific neurons and synapses.
“But it also has great potential to be useful in other settings, like cancer biology where researchers aim to understand the activity of particular cells in the context of the wider tumour.”
The researchers partnered with the electron microscopy team at the Crick, and also worked with synchrotron X-ray teams at the Diamond Light Source in Oxfordshire, The European Synchrotron in France and the Paul Scherrer Institut in Switzerland.
The team will continue this research, using this imaging approach to uncover more detail about the olfactory bulb as well as working to further improve the technique.
About this brain mapping research news
Author: Alice Deeley
Source: Francis Crick Institute
Contact: Alice Deeley – Francis Crick Institute
Image: The image is credited to Carles Bosch
Original Research: Open access.
“Functional and multiscale 3D structural investigation of brain tissue through correlative in vivo physiology, synchrotron microtomography and volume electron microscopy” by Carles Bosch et al. Nature Communications
Abstract
Functional and multiscale 3D structural investigation of brain tissue through correlative in vivo physiology, synchrotron microtomography and volume electron microscopy
Understanding the function of biological tissues requires a coordinated study of physiology and structure, exploring volumes that contain complete functional units at a detail that resolves the relevant features.
Here, we introduce an approach to address this challenge: Mouse brain tissue sections containing a region where function was recorded using in vivo 2-photon calcium imaging were stained, dehydrated, resin-embedded and imaged with synchrotron X-ray computed tomography with propagation-based phase contrast (SXRT).
SXRT provided context at subcellular detail, and could be followed by targeted acquisition of multiple volumes using serial block-face electron microscopy (SBEM).
In the olfactory bulb, combining SXRT and SBEM enabled disambiguation of in vivo-assigned regions of interest. In the hippocampus, we found that superficial pyramidal neurons in CA1a displayed a larger density of spine apparati than deeper ones.
Altogether, this approach can enable a functional and structural investigation of subcellular features in the context of cells and tissues.






