Summary: Scientists developed a computer simulation that models neuron growth in the brain, which could support advancements in neurodegenerative disease research. The simulation accurately replicated real neuron growth patterns in the hippocampus, a brain region key to memory.
Built using BioDynaMo software, the model uses Approximate Bayesian Computation to closely match real-life neuron data, improving its precision. Though the simulation has shown success with specific neuron types, it may need further adjustments for broader applications.
Researchers hope this technology can lead to breakthroughs in understanding and treating conditions like Alzheimer’s. The model’s success points to the potential of digital simulations in enhancing brain research.
Key Facts
- The simulation accurately mimicked neuron growth in the hippocampus.
- The model uses Approximate Bayesian Computation for fine-tuning realism.
- Built on BioDynaMo software, the tool aids in diverse biological simulations.
Source: University of Surrey
A new computer simulation of how our brains develop and grow neurons has been built by scientists from the University of Surrey.
Along with improving our understanding of how the brain works, researchers hope that the models will contribute to neurodegenerative disease research and, someday, stem cell research that helps regenerate brain tissue.

The research team used a technique called Approximate Bayesian Computation (ABC), which helps fine-tune the model by comparing the simulation with real neuron growth. This process ensures that the artificial brain accurately reflects how neurons grow and form connections in real life.
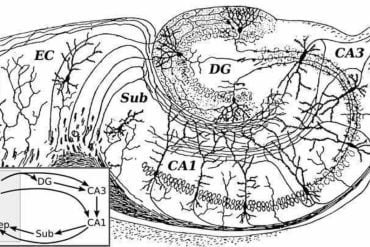
The simulation was tested using neurons from the hippocampus—a critical region of the brain involved in memory retention. The team found that their system successfully mimicked the growth patterns of real hippocampal neurons, showing the potential of this technology to simulate brain development in fine detail.
Dr Roman Bauer from the University of Surrey’s School of Computer Science and Electronic Engineering said:
“How our brain works is still one of the greatest mysteries in science. With this simulation, and the rapid advancements in artificial intelligence, we’re getting closer to understanding how neurons grow and communicate.
“We hope that one day this work could lead to better treatments for devastating diseases like Alzheimer’s or Parkinson’s—changing lives for millions.”
The accuracy of the model is closely tied to the quality of the data used to calibrate it. If the real-life neuron data is limited or incomplete, the precision of the simulation may decrease.
While the current model has shown impressive results in replicating the growth of specific neurons, such as hippocampal pyramidal cells, further adjustments may be needed to accurately simulate other types of neurons or regions of the brain.
The computer simulation is built from the BioDynaMo software, which Dr Bauer co-developed. The software supports scientists to easily create, run, and visualise multi-dimensional agent-based simulations, be they biological, sociological, ecological or financial.
About this computational neuroscience research news
Author: Dalitso Njolinjo
Source: University of Surrey
Contact: Dalitso Njolinjo – University of Surrey
Image: The image is credited to Neuroscience News
Original Research: Open access.
“Calibration of stochastic, agent-based neuron growth models with approximate Bayesian computation” by Roman Bauer et al. Journal of Mathematical Biology
Abstract
Calibration of stochastic, agent-based neuron growth models with approximate Bayesian computation
Understanding how genetically encoded rules drive and guide complex neuronal growth processes is essential to comprehending the brain’s architecture, and agent-based models (ABMs) offer a powerful simulation approach to further develop this understanding. However, accurately calibrating these models remains a challenge.
Here, we present a novel application of Approximate Bayesian Computation (ABC) to address this issue. ABMs are based on parametrized stochastic rules that describe the time evolution of small components–the so-called agents–discretizing the system, leading to stochastic simulations that require appropriate treatment.
Mathematically, the calibration defines a stochastic inverse problem. We propose to address it in a Bayesian setting using ABC. We facilitate the repeated comparison between data and simulations by quantifying the morphological information of single neurons with so-called morphometrics and resort to statistical distances to measure discrepancies between populations thereof.
We conduct experiments on synthetic as well as experimental data. We find that ABC utilizing Sequential Monte Carlo sampling and the Wasserstein distance finds accurate posterior parameter distributions for representative ABMs. We further demonstrate that these ABMs capture specific features of pyramidal cells of the hippocampus (CA1).
Overall, this work establishes a robust framework for calibrating agent-based neuronal growth models and opens the door for future investigations using Bayesian techniques for model building, verification, and adequacy assessment.