Melanoma-like cells in tadpoles may mimic variability in human responses to cancer stimuli.
Uncle Joe smokes a pack a day, drinks like a fish and lives to a ripe old age. His brother, leading a similar lifestyle, succumbs to cancer at age 55. Why do some individuals develop certain diseases or disorders while others do not? In newly reported research that could help provide answers, scientists at Tufts University, in collaboration with the University of Florida, have developed a novel approach that uses artificial intelligence to illuminate cellular processes and suggest possible targets to correct aberrations.
The findings, published Oct. 6 in Science Signaling online in advance of print, are believed to mark the first time artificial intelligence has been used to discover a molecular model that explains why some groups of cells deviate from normal development during embryogenesis, said senior author Michael Levin, Ph.D., the Vannevar Bush Professor of Biology at Tufts and director of the Tufts Center for Regenerative and Developmental Biology.
The paper builds on the center’s earlier studies to understand development and metastasis of melanoma-like cells in tadpoles as well as work applying artificial intelligence to help explain planarian regeneration. The new findings, Levin said, indicate that “our methodology can be taken well beyond simple organisms and applied to the physiology of cell behavior in vertebrates.”
For the Science Signaling work, the researchers applied a type of artificial intelligence called evolutionary computation to pinpoint the molecular mechanisms underlying earlier research in which they induced normal pigment cells in embryonic Xenopus laevis frogs to metastasize. Researchers used a series of drugs to disrupt the cells’ normal bioelectrical and serotonergic signaling at a crucial stage of development. Even in the absence of DNA damage or exposure to carcinogens, the pigment cells of the affected embryos acquired bizarre, branch-like shapes and developed other melanoma-like characteristics, proliferating uncontrollably and invading the frogs’ internal organs.
Depending on which protein in the bioelectric pathway was tweaked, only a certain percentage of the frogs developed melanoma, while the rest remained healthy. “There’s randomness to this process. It doesn’t have the same result in all animals exposed to precisely the same agent, which may mimic the variability in human responses to cancer-inducing stimuli,” said Levin.
Furthermore, the tadpoles that did develop melanoma developed it in every pigment cell–each frog was either 100 percent metastatic or completely normal. Essentially, said Levin, all pigment cells in a tadpole are part of a single coin, which either flips heads (normal) or tails (cancerous). “Metastasis appears to be a group dynamic rather than a single-cell decision,” he said.
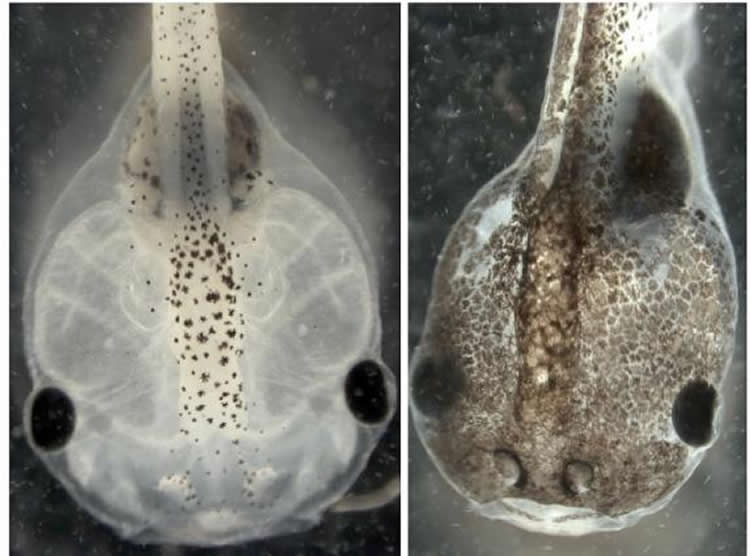
The recent research applied evolutionary computation to understand this complex cell behavior.
Maria Lobikin, Ph.D., recent doctoral graduate from the Levin laboratory and first author on the Science Signaling paper, first identified the building blocks – receptors, hormones and other signaling proteins – of the serotonergic signaling pathway that regulated the melanoma-like cells’ behavior. Then, the team applied artificial intelligence which mimicked evolution to generate a chemical signaling network in a “virtual embryo” that exhibited the same behavior that the researchers observed in their experiments with real tadpoles.
Like biological evolution, evolutionary computation does not randomly or exhaustively test each possibility, but instead uses incremental improvement and selection to rapidly converge on a solution. “The artificial intelligence system evolved a pathway that correctly explains all the existing and very puzzling data. Best of all, it also made correct predictions on data it had never seen,” Levin said.
The knowledge gleaned about these molecular signaling pathways has implications for finding new treatments and targets for tumor prevention and better understanding many other seemingly random decisions made by cells in living organisms. When enough data are available, Levin said, researchers could use this approach to develop a system to help doctors understand patients’ individual genetic responses to treatments as well as environmental factors that cause cancer.
In addition to Levin and Lobikin, paper authors were Douglas J. Blackiston and Elizabeth Tkachenko of the Department of Biology and Center for Regenerative and Developmental Biology, Tufts University; Daniel Lobo, formerly of the Levin laboratory and now at the University of Maryland in Baltimore; and Christopher J. Martyniuk of the Center for Environmental and Human Toxicology and Department of Physiological Sciences, UF Genetics Institute, University of Florida.
Funding: This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation. Computation used a cluster computer awarded by Silicon Mechanics and the Campus Champion Allocation for Tufts University TG-TRA 130003 at the Extreme Science and Engineering Discovery Environment, which is supported by NSF grant ACI-1053575.
Source: Kim Thruler – Tufts University
Image Source: The image is credited to Levin Lab/Tufts University
Original Research: Abstract for “Serotonergic regulation of melanocyte conversion: A bioelectrically regulated network for stochastic all-or-none hyperpigmentation” by Maria Lobikin, Daniel Lobo, Douglas J. Blackiston, Christopher J. Martyniuk, Elizabeth Tkachenko, and Michael Levin in Science Signaling. Published online October 6 2015 doi:10.1126/scisignal.aac6609