Summary: Researchers have created computer models of the neural connections in both health brains and brains of people suffering from Parkinson’s disease.
Source: Frontiers.
A computational study models the strength of basal ganglia connections between healthy and Parkinsonian brains.
Some 10 million people worldwide suffer from Parkinson’s disease — a debilitating condition that causes degeneration of brain nerve cells that control movement. The exact reasons for this degeneration remain unknown. A study published in open-access journal Frontiers in Computational Neuroscience uses a new approach to model the strength of connections within the brain’s basal ganglia. Determining how these differ between healthy and Parkinsonian patients could help scientists understand why individual brains malfunction — and lead to customized therapies specific to the particular pattern of neural degeneration in an individual Parkinson’s sufferer.
Basal ganglia: malfunction causes Parkinson’s
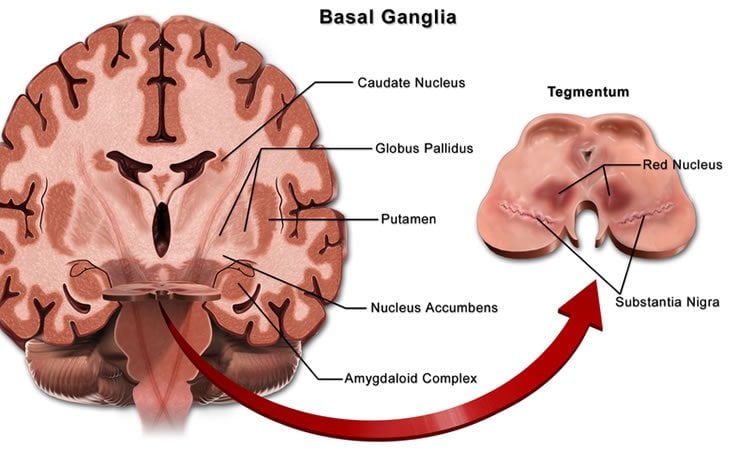
The basal ganglia are a system of neuron clusters, or nuclei, connected to each other and to many other parts of the brain. These ganglia are associated with a number of functions including control of voluntary movements, procedural learning, and habit formation, and their dysfunction leads to neurological disorders such as Parkinson’s disease and Tourette’s syndrome.
“The brain is a complicated organ with many interconnecting systems. At its center, the basal ganglia is one of its most interesting and interconnected regions. Improved experimental methods are now enabling us to discover more about it, revealing the great complexity of the basal ganglia,” says Abigail Morrison of the Institute of Neuroscience and Medicine, Jülich Research Center, Germany.
Every brain is different
Scientists can examine physical connections within the basal ganglia, but many knowledge gaps remain about their strength.
“We need to understand how the strength of connections in the brain’s basal ganglia change when someone develops Parkinson’s disease,” says Morrison. “It’s also important to remember that every brain is different — what is true for one Parkinsonian patient might not be true for another.”
To understand what’s going on in an individual brain, the team devised a way to model this variability. They hope this can be used to develop a computational approach that is complementary to clinical practice. This could lead to the creation of customized therapies, specific to the particular pattern of neural degeneration in an individual Parkinson’s sufferer.
Genetic algorithm helps understand neural connections
There are two main obstacles to accurate modelling of the basal ganglia: the lack of reliable data describing the strength of these connections, and the large variability between subjects. To take these into account, the team decided to generate the parameters for their basal ganglia model using a genetic algorithm.
Genetic algorithms are based on natural selection processes that mimic evolution. In nature, a bird with a harder beak has an advantage foraging for insects in the bark of a tree, and so has a higher probability of reproducing. After many generations, this process can ultimately result in a woodpecker. Analogously, in a genetic algorithm, large numbers of potential solutions are generated, combined, and adapted, until a desired genetic product is reached.
“We turned to the genetic algorithm to generate connections between the nuclei and their strengths computationally. This allowed us to make lots of different network configurations that represent different possible brain connections in individuals,” explains co-author Jyotika Bahuguna.
The team used electrophysiological data from rat brains as criteria for healthy and Parkinsonian conditions. The genetic algorithm generated more than 1,000 possible configurations for the activity occurring in each condition, which were then analyzed for both groups.
“The approach is similar to a weather forecasting technique where predictions for a variety of weather systems are made using different initial configurations of weather conditions,” explains Morrison.
A tool to understand Parkinson’s progression?
The team found a broad overlap between the strength of individual neural connections in healthy and Parkinsonian brains. However, when they looked at the global network activity in response to stimulus, it was easy to determine whether a network configuration was healthy or Parkinsonian. A detailed analysis of these networks, especially the ones which lie on the boundaries between healthy and diseased, might shed light on how a brain transitions from a healthy to a Parkinsonian state. More importantly, it could help identify therapeutically feasible options that would allow transition from a Parkinsonian state back to a healthy state.
“It is important to realize that Parkinson’s disease is dynamic. We have a stylized picture of the effects of neurodegeneration, but we need to remember that the patterns of degeneration are variable and are different in each individual,” concludes Morrison.
“Future clinical approaches incorporating computational methods could incorporate the variability present among the patients. This would allow treatment to be customized to compensate for the specific pattern of degeneration exhibited by a patient, to try to restore healthy dynamics.”
Funding: The Initiative and Networking Fund of the Helmholtz Association, German Research Foundation, EuroSPIN, Erasmus Mundus Joint Doctorate Program supported this research.
Source: Emma Duncan – Frontiers
Image Source: NeuroscienceNews.com image is credited to Bruce Blaus and is licensed CC BY 3.0.
Original Research: Full open access research for “Homologous Basal Ganglia Network Models in Physiological and Parkinsonian Conditions” by Jyotika Bahuguna, Tom Tetzlaff, Arvind Kumar, Jeanette Hellgren Kotaleski and Abigail Morrison in Frontiers in Computational Neuroscience. Published online August 22 2017 doi:10.3389/fncom.2017.00079
[cbtabs][cbtab title=”MLA”]Frontiers “Modeling Brain Connections to Understand Parkinson’s Disease.” NeuroscienceNews. NeuroscienceNews, 27 September 2017.
<https://neurosciencenews.com/parkinsons-brain-connections-7601/>.[/cbtab][cbtab title=”APA”]Frontiers (2017, September 27). Modeling Brain Connections to Understand Parkinson’s Disease. NeuroscienceNews. Retrieved September 27, 2017 from https://neurosciencenews.com/parkinsons-brain-connections-7601/[/cbtab][cbtab title=”Chicago”]Frontiers “Modeling Brain Connections to Understand Parkinson’s Disease.” https://neurosciencenews.com/parkinsons-brain-connections-7601/ (accessed September 27, 2017).[/cbtab][/cbtabs]
Abstract
Maternal anxiety predicts attentional bias towards threat in infancy
The classical model of basal ganglia has been refined in recent years with discoveries of subpopulations within a nucleus and previously unknown projections. One such discovery is the presence of subpopulations of arkypallidal and prototypical neurons in external globus pallidus, which was previously considered to be a primarily homogeneous nucleus. Developing a computational model of these multiple interconnected nuclei is challenging, because the strengths of the connections are largely unknown. We therefore use a genetic algorithm to search for the unknown connectivity parameters in a firing rate model. We apply a binary cost function derived from empirical firing rate and phase relationship data for the physiological and Parkinsonian conditions. Our approach generates ensembles of over 1,000 configurations, or homologies, for each condition, with broad distributions for many of the parameter values and overlap between the two conditions. However, the resulting effective weights of connections from or to prototypical and arkypallidal neurons are consistent with the experimental data. We investigate the significance of the weight variability by manipulating the parameters individually and cumulatively, and conclude that the correlation observed between the parameters is necessary for generating the dynamics of the two conditions. We then investigate the response of the networks to a transient cortical stimulus, and demonstrate that networks classified as physiological effectively suppress activity in the internal globus pallidus, and are not susceptible to oscillations, whereas parkinsonian networks show the opposite tendency. Thus, we conclude that the rates and phase relationships observed in the globus pallidus are predictive of experimentally observed higher level dynamical features of the physiological and parkinsonian basal ganglia, and that the multiplicity of solutions generated by our method may well be indicative of a natural diversity in basal ganglia networks. We propose that our approach of generating and analyzing an ensemble of multiple solutions to an underdetermined network model provides greater confidence in its predictions than those derived from a unique solution, and that projecting such homologous networks on a lower dimensional space of sensibly chosen dynamical features gives a better chance than a purely structural analysis at understanding complex pathologies such as Parkinson’s disease.
“Homologous Basal Ganglia Network Models in Physiological and Parkinsonian Conditions” by Jyotika Bahuguna, Tom Tetzlaff, Arvind Kumar, Jeanette Hellgren Kotaleski and Abigail Morrison in Frontiers in Computational Neuroscience. Published online August 22 2017 doi:10.3389/fncom.2017.00079