Summary: A new study explores how neural activity influences CREB dynamics.
Source: Osaka University.
Neuronal activity mediates the formation of neuronal circuits in the cerebral cortex. These processes are regulated by the transcription factor CREB, which regulates gene expression in neuronal activity-dependent processes. Neuronal activity enhances CREB-mediated transcription but the mechanisms remain unclear. CREB binds to a cAMP response element (CRE) in the promoter region of its target genes. Assembly and disassembly of CREB-CRE interactions control spatiotemporal gene expression in the nucleus. However, how CREB interacts with CRE in activity-dependent mechanisms is not known.
In a new study from Osaka University, led by Nobuhiko Yamamoto, Hironobu Kitagawa, Noriyuki Sugo and colleagues investigated how neuronal activity influences CREB dynamics. Fluorescent-tagged CREB constructs were expressed in primary cortical neurons and visualized by staining fixed cells. Single CREB molecules were predominantly detected in the nucleus and some were observed at sites where gene transcription was taking place.
The movement of single CREB molecules in living cortical neurons was monitored by real-time imaging. Interestingly, some CREB molecules disappeared almost immediately while a fraction remained in the same location for a longer duration. Using mutant forms of CREB that cannot bind to CRE, the researchers showed that these CREB molecules were interacting with CRE.
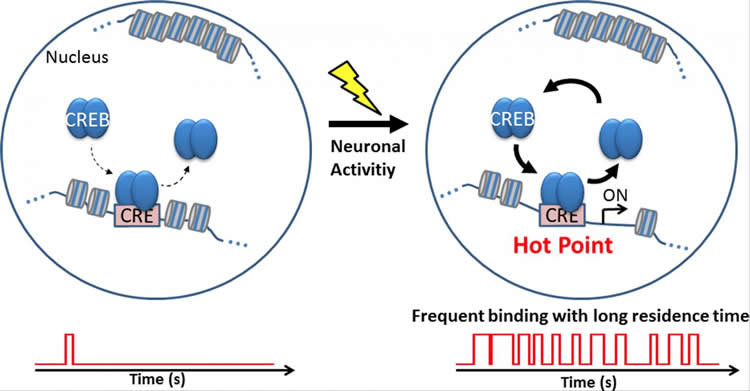
To investigate how neuronal activity affects CREB-CRE interactions and subsequent gene transcription, the authors treated cortical neurons with pharmacological agents that alter neuronal activity. Neuronal activity did not affect the time that CREB resided in the nucleus. Next, the research team investigated whether neuronal activity regulates the spatial distribution of CREB in the nucleus. They stimulated cortical neurons and monitored CREB localization by real-time imaging. Interestingly, the number of locations where CREB resided for long periods increased considerably after neuronal activation. These findings indicated that neuronal activity increases CREB-CRE interactions in the nucleus.
The findings provide novel insights into the regulation of gene expression by neuronal activity. Based on the results of this study, neuronal activity may contribute to CREB-dependent gene expression by increasing the binding of CREB to specific genomic sites.
Funding: This research was funded by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
F31NS081805, NINDS R01NS078294, U54HD083092 to the BCM IDDRC and the Intelligence Advanced Research Projects Activity (IARPA) via Department of Interior/Interior Business Center (DoI/IBC) contract number D16PC00003.
Source: Saori Obayashi – Osaka University
Image Source: NeuroscienceNews.com image is adapted from the CMU press release.
Original Research: Abstract for “Activity-Dependent Dynamics of the Transcription Factor CREB in Cortical Neurons Revealed by Single-Molecule Imaging” by Hironobu Kitagawa, Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Toshio Yanagida and Nobuhiko Yamamoto in Journal of Neuroscience. Published online November 3 2016 doi:10.1523/JNEUROSCI.0943-16.2016
[cbtabs][cbtab title=”MLA”]Osaka University “Novel Insights Into Neuronal Activity-Dependent Gene Expression by CREB.” NeuroscienceNews. NeuroscienceNews, 27 December 2016.
<https://neurosciencenews.com/creb-gene-neural-activity-5820/>.[/cbtab][cbtab title=”APA”]Osaka University (2016, December 27). Novel Insights Into Neuronal Activity-Dependent Gene Expression by CREB. NeuroscienceNew. Retrieved December 27, 2016 from https://neurosciencenews.com/creb-gene-neural-activity-5820/[/cbtab][cbtab title=”Chicago”]Osaka University “Novel Insights Into Neuronal Activity-Dependent Gene Expression by CREB.” https://neurosciencenews.com/creb-gene-neural-activity-5820/ (accessed December 27, 2016).[/cbtab][/cbtabs]
Abstract
Activity-Dependent Dynamics of the Transcription Factor CREB in Cortical Neurons Revealed by Single-Molecule Imaging
Transcriptional regulation is crucial for neuronal activity-dependent processes that govern neuronal circuit formation and synaptic plasticity. An intriguing question is how neuronal activity influences the spatiotemporal interactions between transcription factors and their target sites. Here we investigated the activity dependence of DNA binding and dissociation events of cAMP-response element binding protein (CREB), a principal factor in activity-dependent transcription, in mouse cortical neurons using a single-molecule imaging technique. To visualize CREB at the single-molecule level, fluorescent-tagged CREB in living dissociated cortical neurons was observed by highly inclined and laminated optical sheet (HILO) microscopy. We found that a significant fraction of CREB spots resided in the restricted locations in the nucleus for several seconds (dissociation rate constant: 0.42 s-1). In contrast, two mutant CREBs, which cannot bind to the cAMP-response element (CRE), scarcely exhibited long-term residence. To test the possibility that CREB dynamics depends on neuronal activity, pharmacological treatments and an optogenetic method involving Channelrhodopsin-2 were applied to cultured cortical neurons. Increased neuronal activity did not appear to influence the residence time of CREB spots, but markedly increased the number of restricted locations (hot spots) where CREB spots frequently resided with long residence times (> 1 s). These results suggest that neuronal activity promotes CREB-dependent transcription by increasing the frequency of CREB binding to highly localized genome locations.
SIGNIFICANCE STATEMENT
The transcription factor, CREB (cAMP response element-binding protein) is known to regulate gene expression in neuronal activity-dependent processes. However, its spatiotemporal interactions with the genome remain unknown. Single-molecule imaging in cortical neurons revealed that fluorescent-tagged CREB spots frequently reside at fixed nuclear locations in the time range of several seconds. Neuronal activity had little effect on the CREB residence time, but increased the rapid and frequent reappearance of long-residence CREB spots at the same nuclear locations. Thus, activity-dependent transcription is attributable to frequent binding of CREB to specific genome loci.
“Activity-Dependent Dynamics of the Transcription Factor CREB in Cortical Neurons Revealed by Single-Molecule Imaging” by Hironobu Kitagawa, Noriyuki Sugo, Masatoshi Morimatsu, Yoshiyuki Arai, Toshio Yanagida and Nobuhiko Yamamoto in Journal of Neuroscience. Published online November 3 2016 doi:10.1523/JNEUROSCI.0943-16.2016