Summary: Researchers report looking at neural networks in the brain, rather than just specific regions, could shed new light on why some brain injuries are worse than others.
Source: PLOS.
Considering the brain’s network of activity, rather than just individual regions, could help us understand why some brain injuries are much worse than others, according to a study published PLOS Computational Biology by Maxwell B. Wang, Julia Owen, and Pratik Mukherjee from University of California, San Francisco, and Ashish Raj from Weill Cornell Medicine.
The human brain displays an astonishing range of responses to injury, depending on its location. This is conventionally considered a result of the fact that each brain region has a specific functional role. However, there is increasing evidence that the brain’s regions do not operate in isolation but as a network or ‘connectome.’ Therefore, to understand the effect of injury, we must look not just for localized changes but network-wide changes caused by the disruption of network connections.
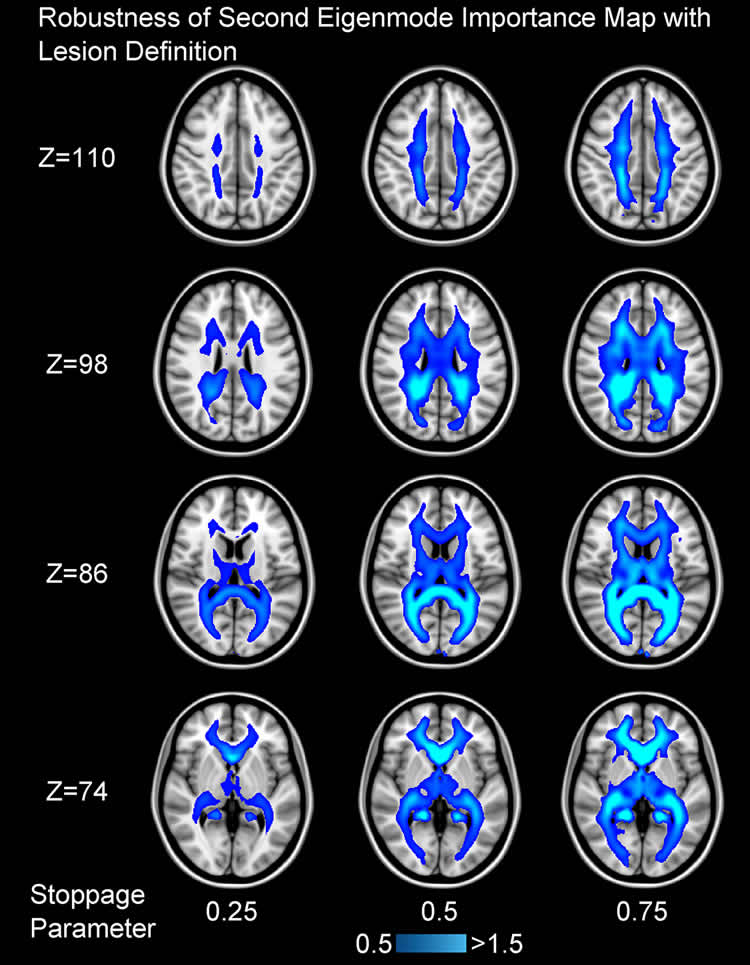
The authors used a combination of Diffusion-Tensor MRI and ideas from graph theory to investigate patterns in the white matter network architecture of the brain, and how these change due to injury. They built on previous work by Dr. Raj, in the Feil Family Brain and Mind Research Institute at Weill Cornell Medicine, and colleagues that showed that simple models of networks capture brain-wide patterns of degeneration as well as normal patterns of brain activity. These patterns can be decomposed into the network’s “eigenmodes,” which may be thought of as sub-networks into which reverberatory or propagating activity gets concentrated, not unlike vibrating modes on a guitar string.
The authors first showed that the most important eigenmodes are persistently found across healthy subjects, and in multiple scans of the same subject. They modelled the effect of brain lesions on eigenmodes and found that the influence of a lesion on network eigenmodes is fairly sensitive to the site of the lesion. The researchers identified white matter fiber pathways that were particularly responsible for the flow of information throughout the brain. They found that the lesions that most influenced network eigenmodes were those at the center of these pathways.
They looked in particular at a rare neurodevelopmental disorder where the corpus callosum, the primary connection between the left and right hemispheres of the brain, is absent. Patients suffering from this disease typically fare much better than those whose corpus callosum is surgically removed, an observation which can be explained using the eigenmode model.
If these findings can be replicated in other scenarios, it could be possible to predict with greater accuracy how future changes to brain structure, such as surgery or the progression of a disease, would result in changes to brain function, allowing physicians to better tailor treatment plans and therapeutic targets.
“This study proposes a method to relate how changes in the brain’s structural architecture can result in the observable deficits we identify with neurological disorders. We hope to extend these methods towards predicting how future perturbations to a patient’s brain, either from the progression of a disease or a novel therapeutic, can affect neurological function,” says senior study author Dr. Raj.
Funding: AR was supported by National Institute of Biomedical Imaging and Bioengineering grant EB022717 and National Institute of Neurological Disorders grant NS092802. PM was supported by Simons Foundation grant SFARI A118077. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Source: Renee Tessman – PLOS
Image Source: NeuroscienceNews.com image is credited to Raj et al./PLOS.
Original Research: Full open access research for “Brain network eigenmodes provide a robust and compact representation of the structural connectome in health and disease” by Maxwell B. Wang, Julia P. Owen, Pratik Mukherjee, and Ashish Raj in PLOS Computational Biology. Published online June 22 2017 doi:10.1371/journal.pcbi.1005550
[cbtabs][cbtab title=”MLA”]PLOS “New Brain Network Model Could Explain Differences in Brain Injuries.” NeuroscienceNews. NeuroscienceNews, 23 June 2017.
<https://neurosciencenews.com/tbi-brain-network-6969/>.[/cbtab][cbtab title=”APA”]PLOS (2017, June 23). New Brain Network Model Could Explain Differences in Brain Injuries. NeuroscienceNew. Retrieved June 23, 2017 from https://neurosciencenews.com/tbi-brain-network-6969/[/cbtab][cbtab title=”Chicago”]PLOS “New Brain Network Model Could Explain Differences in Brain Injuries.” https://neurosciencenews.com/tbi-brain-network-6969/ (accessed June 23, 2017).[/cbtab][/cbtabs]
Abstract
Brain network eigenmodes provide a robust and compact representation of the structural connectome in health and disease
Recent research has demonstrated the use of the structural connectome as a powerful tool to characterize the network architecture of the brain and potentially generate biomarkers for neurologic and psychiatric disorders. In particular, the anatomic embedding of the edges of the cerebral graph have been postulated to elucidate the relative importance of white matter tracts to the overall network connectivity, explaining the varying effects of localized white matter pathology on cognition and behavior. Here, we demonstrate the use of a linear diffusion model to quantify the impact of these perturbations on brain connectivity. We show that the eigenmodes governing the dynamics of this model are strongly conserved between healthy subjects regardless of cortical and sub-cortical parcellations, but show significant, interpretable deviations in improperly developed brains. More specifically, we investigated the effect of agenesis of the corpus callosum (AgCC), one of the most common brain malformations to identify differences in the effect of virtual corpus callosotomies and the neurodevelopmental disorder itself. These findings, including the strong correspondence between regions of highest importance from graph eigenmodes of network diffusion and nexus regions of white matter from edge density imaging, show converging evidence toward understanding the relationship between white matter anatomy and the structural connectome.
“Brain network eigenmodes provide a robust and compact representation of the structural connectome in health and disease” by Maxwell B. Wang, Julia P. Owen, Pratik Mukherjee, and Ashish Raj in PLOS Computational Biology. Published online June 22 2017 doi:10.1371/journal.pcbi.1005550