International groups join forces to find elusive gene variants in largest-ever sample set.
An international group of researchers has identified 11 new genes that offer important new insights into the disease pathways involved in Alzheimer’s disease. The highly collaborative effort involved scanning the DNA of over 74,000 volunteers—the largest genetic analysis yet conducted in Alzheimer’s research—to discover new genetic risk factors linked to late-onset Alzheimer’s disease, the most common form of the disorder.
By confirming or suggesting new processes that may influence Alzheimer’s disease development—such as inflammation and synaptic function—the findings point to possible targets for the development of drugs aimed directly at prevention or delaying disease progression.
Supported in part by the National Institute on Aging (NIA) and other components of the National Institutes of Health, the International Genomic Alzheimer’s Project (IGAP) reported its findings online in Nature Genetics on Oct. 27, 2013. IGAP is comprised of four consortia in the United States and Europe which have been working together since 2011 on genome-wide association studies (GWAS) involving thousands of DNA samples and shared datasets. GWAS are aimed at detecting the subtle gene variants involved in Alzheimer’s and defining how the molecular mechanisms influence disease onset and progression.

“Collaboration among researchers is key to discerning the genetic factors contributing to the risk of developing Alzheimer’s disease,” said Richard J. Hodes, M.D., director of the NIA. “We are tremendously encouraged by the speed and scientific rigor with which IGAP and other genetic consortia are advancing our understanding.”
The search for late-onset Alzheimer’s risk factor genes had taken considerable time, until the development of GWAS and other techniques. Until 2009, only one gene variant, Apolipoprotein E-e4 (APOE-e4), had been identified as a known risk factor. Since then, prior to today’s discovery, the list of known gene risk factors had grown to include other players—PICALM, CLU, CR1, BIN1, MS4A, CD2AP, EPHA1, ABCA7, SORL1 and TREM2.
IGAP’s discovery reported today of 11 new genes strengthens evidence about the involvement of certain pathways in the disease, such as the role of the SORL1 gene in the abnormal accumulation of amyloid protein in the brain, , a hallmark of Alzheimer’s disease. It also offers new gene risk factors that may influence several cell functions, to include the ability of microglial cells to respond to inflammation.
The researchers identified the new genes by analyzing previously studied and newly collected DNA data from 74,076 older volunteers with Alzheimer’s and those free of the disorder from 15 countries. The new genes (HLA-DRB5/HLA0DRB1, PTK2B, SLC24A4-0RING3, DSG2, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2 and CASS4) add to a growing list of gene variants associated with onset and progression of late-onset Alzheimer’s. Researchers will continue to explore the roles played by these genes, to include:
- How SORL1 and CASS4 influence amyloid, and how CASS4 and FERMT2 affect tau, another protein hallmark of Alzheimer’s disease
- How inflammation is influenced by HLA-DRB5/DRB1, INPP5D, MEF2C, CR1 and TREM2
- How SORL1affects lipid transport and endocytosis (or protein sorting within cells)
- How MEF2C and PTK2B influence synaptic function in the hippocampus, a brain region important to learning and memory
- How CASS4, CELF1, NME8 and INPP5 affect brain cell function
The study also brought to light another 13 variants that merit further analysis.
“Interestingly, we found that several of these newly identified genes are implicated in a number of pathways,” said Gerard Schellenberg, Ph.D., University of Pennsylvania School of Medicine, Philadelphia, who directs one of the major IGAP consortia. “Alzheimer’s is a complex disorder, and more study is needed to determine the relative role each of these genetic factors may play. I look forward to our continued collaboration to find out more about these—and perhaps other—genes.”
Schellenberg heads the Alzheimer’s Disease Genetics Consortium (ADGC), one of the four founding partners of IGAP. The ADGC is a collaborative body established and funded by the NIA with the goal of identifying genetic variants associated with risk for Alzheimer’s. Schellenberg noted that the study was made possible by the research infrastructures established and supported by the NIA over many years, including 29 Alzheimer’s Disease Centers, the National Alzheimer’s Coordinating Center, the NIA Genetics of Alzheimer’s Disease Data Storage Site, the Late-onset Alzheimer’s Disease Family Study, and the National Cell Repository for Alzheimer’s Disease. These endeavors collect, store and make available to qualified researchers DNA samples, datasets containing biomedical and demographic information about participants, and genetic analysis data.
The other three founding partners of IGAP are: The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) led by Sudha Seshadri at Boston University and supported in part by NIH (including NIH-supported databases from the AGES-Reykjavik Study and the Atherosclerosis Risk in Communities Study); the European Alzheimer’s Disease Initiative (EADI) led by Philippe Amouyel of Lille University, France; and Genetic and Environmental Research in Alzheimer’s Disease (GERAD) led by Julie Williams of Cardiff University, Wales.
Notes about this Alzheimer’s disease and neurogenetics research
The efforts were also supported by the Alzheimer’s Association and an extensive number of international governmental, private, and public research groups.
Contact: Peggy Vaughn – NIH/NIA
Source: NIH/NIA press release
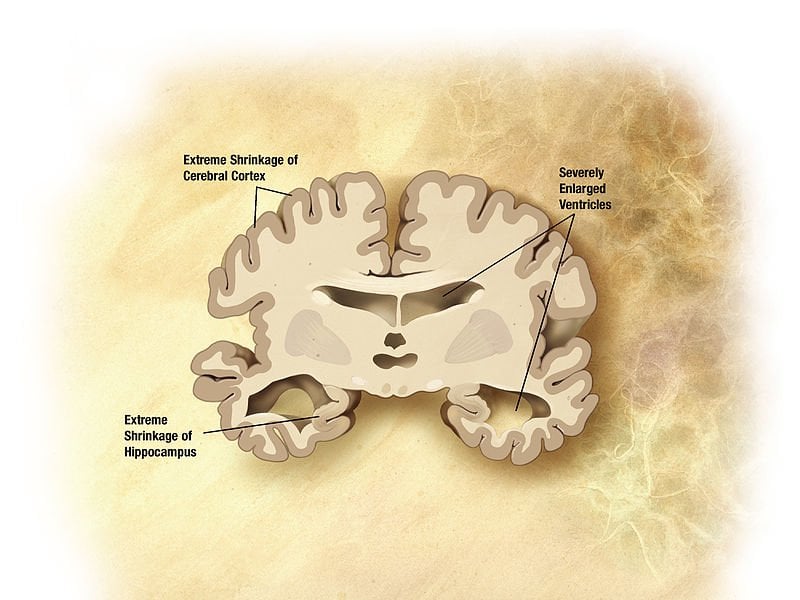
Image Source: The Alzheimer’s brain image is adapted from an NIA/NIH image and is in the public domain.
Original Research: Abstract for “Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease” by Jean-Charles Lambert, Carla A Ibrahim-Verbaas, Denise Harold, Adam C Naj, Rebecca Sims, Céline Bellenguez, Gyungah Jun, Anita L DeStefano, Joshua C Bis, Gary W Beecham, Benjamin Grenier-Boley, Giancarlo Russo, Tricia A Thornton-Wells, Nicola Jones, Albert V Smith, Vincent Chouraki, Charlene Thomas, M Arfan Ikram, Diana Zelenika, Badri N Vardarajan, Yoichiro Kamatani, Chiao-Feng Lin, Amy Gerrish, Helena Schmidt, Brian Kunkle, Melanie L Dunstan, Agustin Ruiz, Marie-Thérèse Bihoreau, Seung-Hoan Choi, Christiane Reitz, Florence Pasquier, Paul Hollingworth, Alfredo Ramirez, Olivier Hanon, Annette L Fitzpatrick, Joseph D Buxbaum, Dominique Campion, Paul K Crane, Clinton Baldwin, Tim Becker, Vilmundur Gudnason, Carlos Cruchaga, David Craig, Najaf Amin, Claudine Berr, Oscar L Lopez, Philip L De Jager, Vincent Deramecourt, Janet A Johnston, Denis Evans, Simon Lovestone, Luc Letenneur, Francisco J Morón, David C Rubinsztein Gudny Eiriksdottir, Kristel Sleegers, Alison M Goate, Nathalie Fiévet, Matthew J Huentelman, Michael Gill, Kristelle Brown, M Ilyas Kamboh, Lina Keller, Pascale Barberger-Gateau, Bernadette McGuinness, Eric B Larson, Robert Green, Amanda J Myers, Carole Dufouil, Stephen Todd, David Wallon, Seth Love, Ekaterina Rogaeva, John Gallacher, Peter St George-Hyslop, Jordi Clarimon, Alberto Lleo, Anthony Bayer, Debby W Tsuang, Lei Yu, Magda Tsolaki, Paola Bossù, Gianfranco Spalletta, Petroula Proitsi, John Collinge, Sandro Sorbi, Florentino Sanchez-Garcia, Nick C Fox, John Hardy, Maria Candida Deniz Naranjo, Paolo Bosco, Robert Clarke, Carol Brayne, Daniela Galimberti, Michelangelo Mancuso, Fiona Matthews, Susanne Moebus, Patrizia Mecocci, Maria Del Zompo, Wolfgang Maier, Harald Hampel, Alberto Pilotto, Maria Bullido, Francesco Panza, Paolo Caffarra, Benedetta Nacmias, John R Gilbert, Manuel Mayhaus, Lars Lannfelt, Hakon Hakonarson, Sabrina Pichler, Minerva M Carrasquillo, Martin Ingelsson, Duane Beekly, Victoria Alvarez, Fanggeng Zou, Otto Valladares, Steven G Younkin, Eliecer Coto, Kara L Hamilton-Nelson, Wei Gu, Cristina Razquin, Pau Pastor, Ignacio Mateo, Michael J Owen, Kelley M Faber, Palmi V Jonsson, Onofre Combarros, Michael C O’Donovan, Laura B Cantwell, Hilkka Soininen, Deborah Blacker, Simon Mead, Thomas H Mosley, David A Bennett, Tamara B Harris, Laura Fratiglioni, Clive Holmes, Renee F A G de Bruijn, Peter Passmore, Thomas J Montine, Karolien Bettens, Jerome I Rotter, Alexis Brice, Kevin Morgan, Tatiana M Foroud, Walter A Kukull, Didier Hannequin, John F Powell, Michael A Nalls, Karen Ritchie, Kathryn L Lunetta, John S K Kauwe, Eric Boerwinkle, Matthias Riemenschneider, Mercè Boada, Mikko Hiltunen, Eden R Martin, Reinhold Schmidt, Dan Rujescu, Li-San Wang, Jean-François Dartigues, Richard Mayeux, Christophe Tzourio, Albert Hofman, Markus M Nöthen, Caroline Graff, Bruce M Psaty, Lesley Jones, Jonathan L Haines, Peter A Holmans, Mark Lathrop, Margaret A Pericak-Vance, Lenore J Launer, Lindsay A Farrer, Cornelia M van Duijn, Christine Van Broeckhoven, Valentina Moskvina, Sudha Seshadri, Julie Williams, Gerard D Schellenberg and Philippe Amouyel in Nature Genetics. Published online October 27 2013 doi:10.1038/ng.2802