Responding appropriately to the smell of food or the scent of danger can mean life or death to a fruit fly, and dedicated circuits in the insect’s brain are in place to make sure the fly gets it right.
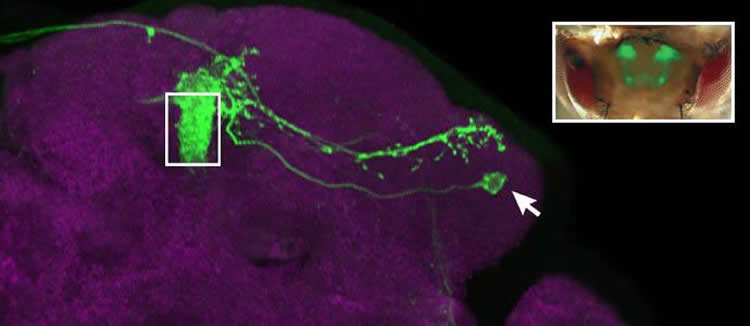
In studies designed to better understand how the brain processes information, scientists led by Cold Spring Harbor Laboratory (CSHL) Associate Professor Glenn Turner have identified an important component in these circuits: the point at which incoming sensory information begins to be transformed into a neural signal that instructs a fly’s response. The cells, called mushroom body output neurons (MBONs), appear to distill nuanced information about an odor into clear instructions: approach or flee.
By genetically labeling and following the activity of the same MBONs in multiple flies (there are precisely 34 in each brain, situated in known locations), the scientists found that each cell had a characteristic response pattern in each individual. The pattern, in other words, differed between flies. This suggests that MBONs may underlie individual odor preferences that develop as flies learn to associate smells with positive or negative experiences.
Mushroom body output neurons were discovered last year by Turner’s collaborators, Gerald Rubin and Yoshinori Aso, at the Janelia Research Campus of the Howard Hughes Medical Institute. As their name implies, the cells are located in the mushroom body, a part of the fruit fly brain critical for learned responses to sensory stimuli. The mushroom body itself is composed of roughly 2000 neurons that transmit information about smells that have been detected by the fly’s sensory neurons – and on the receiving end of these transmissions are just 34 MBONs. A fly’s behavioral response to an odor depends on the message that these output neurons relay to neurons farther along the circuit.
Toshihide Hige, a postdoctoral researcher in Turner’s lab, measured how MBONs responded to 10 different smells–an assortment of potentially enticing food smells, like yeast and vinegar; aversive smells such as citronella; and more neutral aromas. Responses varied greatly between cells, with each cell most strongly attuned to certain aromas.
When the team analyzed the cells’ responses, some patterns emerged. In general, food smells elicited response patterns that were distinct from those elicited by repellant odors, suggesting that although mushroom body output cells probably don’t identify specific odors, they may communicate each odor’s most essential quality–whether it is “good” or “bad.” This can be the basis for action – to pursue the odor or fly away from it.
In each fly, pairs of matched MBONs on opposite hemispheres of the fly brain, which could be identified both genetically and anatomically, shared the same response patterns. But when the scientists examined the corresponding cells in a different fly, they exhibited a whole new pattern of activity. That suggested that MBON sensitivities had been established through the past experiences of the individual flies being examined.
“As a fly lives its life and encounters a bunch of different odors, that olfactory experience may induce some plasticity in the circuit,” Turner says, explaining that this could allow a fly’s brain to adjust olfactory preferences.
To explore this idea, the team repeated their experiments, this time in flies that lacked a memory-related gene called rutabaga. MBONs in these learning-impaired flies still fired in response to odors, but the individuality of each cell’s response was lost. Activity patterns of each cell looked more or less the same in different animals. “It looks like plasticity — the result of particular experiences — is driving each of these output neurons to their own particular response properties, which are different for each individual fly,” Turner says.
These fly-to-fly differences in cell signaling reflect differences in olfactory preferences that are present not just in fruit flies, but also in humans, Turner notes. “We all have slightly different olfactory perceptions. I kind of hate the smell of durians [a tropical fruit], but my wife really likes the smell of durians. One person’s perfume is another person’s horrible stench.”
Funding: This work was supported by the National Institutes of Health, the Japan Society for the Promotion of Science, and the Uehara Memorial Foundation.
Source: Peter Tarr – CSHL
Image Source: The image is credited to Turner Lab, CSHL
Original Research: Abstract for “Plasticity-driven individualization of olfactory coding in mushroom body output neurons” by Toshihide Hige, Yoshinori Aso, Gerald M. Rubin and Glenn C. Turner in Nature. Published online September 30 2015 doi:10.1038/nature15396
Abstract
Plasticity-driven individualization of olfactory coding in mushroom body output neurons
Although all sensory circuits ascend to higher brain areas where stimuli are represented in sparse, stimulus-specific activity patterns, relatively little is known about sensory coding on the descending side of neural circuits, as a network converges. In insects, mushroom bodies have been an important model system for studying sparse coding in the olfactory system1, 2, 3, where this format is important for accurate memory formation4, 5, 6. In Drosophila, it has recently been shown that the 2,000 Kenyon cells of the mushroom body converge onto a population of only 34 mushroom body output neurons (MBONs), which fall into 21 anatomically distinct cell types7, 8. Here we provide the first, to our knowledge, comprehensive view of olfactory representations at the fourth layer of the circuit, where we find a clear transition in the principles of sensory coding. We show that MBON tuning curves are highly correlated with one another. This is in sharp contrast to the process of progressive decorrelation of tuning in the earlier layers of the circuit2, 9. Instead, at the population level, odour representations are reformatted so that positive and negative correlations arise between representations of different odours. At the single-cell level, we show that uniquely identifiable MBONs display profoundly different tuning across different animals, but that tuning of the same neuron across the two hemispheres of an individual fly was nearly identical. Thus, individualized coordination of tuning arises at this level of the olfactory circuit. Furthermore, we find that this individualization is an active process that requires a learning-related gene, rutabaga. Ultimately, neural circuits have to flexibly map highly stimulus-specific information in sparse layers onto a limited number of different motor outputs. The reformatting of sensory representations we observe here may mark the beginning of this sensory-motor transition in the olfactory system.
“Plasticity-driven individualization of olfactory coding in mushroom body output neurons” by Toshihide Hige, Yoshinori Aso, Gerald M. Rubin and Glenn C. Turner in Nature. Published online September 30 2015 doi:10.1038/nature15396






