Summary: Researchers from UT Southwestern’s Cryo-EM Facility have determined the 3D anatomical structure of the TRPML1 ion channel. The ion channel has been implicated in mucolipidosis type IV, a rare, genetic neurodegenerative disease.
Source: UT Southwestern.
UT Southwestern Medical Center researchers today published a 3-D atomic structure of the ion channel found in mammals that is implicated in a rare, inherited neurodegenerative disease in humans. The work marks the first such structure determined using the university’s $17 million cryo-electron microscopy (cryo-EM) facility that opened last year.
In cryo-EM, samples are rapidly frozen to prevent the formation of damaging ice crystals and then viewed at minus 321 degrees Fahrenheit (cryogenic temperatures). UT Southwestern’s facility – which runs round-the-clock – is one of the world’s top facilities for cryo-EM structural biology.
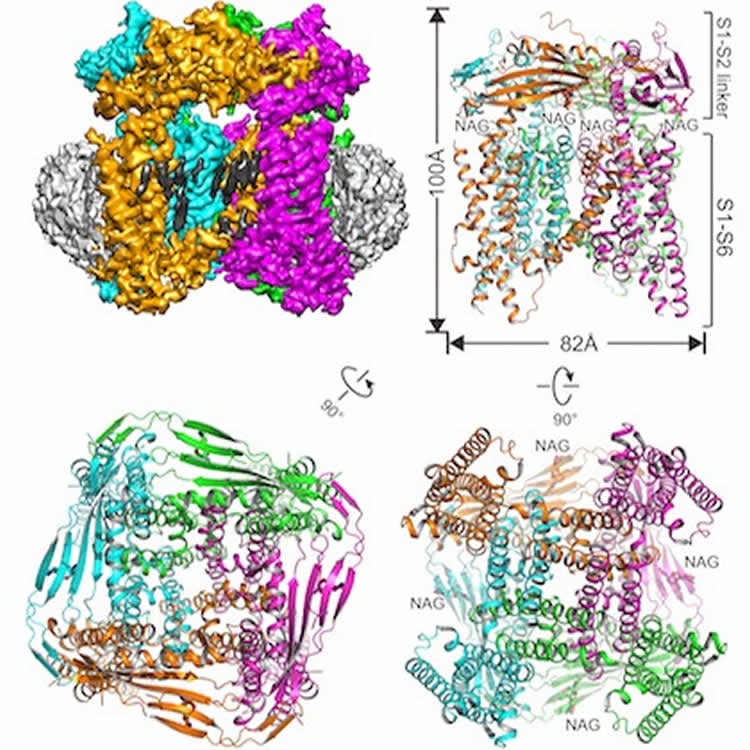
The interdisciplinary team solved the structure of the mouse TRPML1 (transient receptor potential mucolipin 1) ion channel.
Their study, published online by Nature, comes just a week after three developers of cryo-EM technology were recognized with the 2017 Nobel Prize in Chemistry for their work on the instruments and methodologies that sparked the “resolution revolution.” Cryo-EM enables atomic-level views of proteins that resist the crystallization necessary for traditional X-ray crystallography.
“Functioning ion channels are needed for the proper movement of electrically charged particles – ions – in and out of cells and organelles to run cellular processes,” said Dr. Youxing Jiang, Professor of Physiology and Biophysics, an Investigator in the Howard Hughes Medical Institute (HHMI), and co-corresponding author of the study.
The TRPML1 channel, which regulates the flow of calcium ions, is found in every mammal. The channel sits in the membrane of organelles inside cells called lysosomes, which contain enzymes that aid in cellular recycling by breaking down large molecules.
About 50 lysosomal storage diseases (LSDs) have been identified in humans, including one class of LSDs caused by loss-of-function mutations in genes governing the TRPML1 channels. This LSD, called mucolipidosis type IV, is marked by delayed development of mental and motor skills and vision impairment, according to the National Institutes of Health.
“Due to its link to that class of lysosomal storage diseases, TRPML1 has been a potential target for small-molecule therapeutics and several potential agonists (channel openers) have been developed,” said Dr. Jiang, a W.W. Caruth, Jr. Scholar in Biomedical Research. Determination of TRPML1’s structure could aid in the search for treatments for mucolipidosis type IV, he said.
UT Southwestern’s cryo-EM facility houses three high-tech instruments, including a 12½-foot-tall, 2-ton Titan Krios that shoots a high-powered beam through each sample while a special camera captures images of the scattered electrons that result. A robotic arm inside the machine can hold and precisely move a dozen flash-frozen samples in an automated manner so that thousands of images can be recorded, processed via computers, and interpreted to generate 3-D images for study, the researchers explained.
Another distinction in this study is the successful use of a relatively new sample preparation technique: embedding the protein of interest in a nanodisc structure made from lipids and other biological materials. Lead author Dr. Qingfeng Chen, a postdoctoral researcher in the Jiang laboratory, created the sample.
“For a long time, detergent has been used to extract proteins from membranes for study. People have suggested that detergent might change the protein structure from its native state,” said Dr. Xiaochen Bai, an Assistant Professor of Biophysics and Cell Biology and the study’s second corresponding author. “Membrane proteins, such as those we studied, are usually wrapped in lipids. Nanodiscs are used to provide a native environment for the protein sample.”
Dr. Bai specializes in the sophisticated software used to reveal the organization of individual atoms within each particle. He was recruited to UT Southwestern as a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar and a Virginia Murchison Linthicum Scholar in Medical Research after five years of training at the MRC Laboratory of Molecular Biology in Cambridge, U.K.
Working on a team developing what has become one of the top software programs for processing the image data generated by the electron microscopy, Dr. Bai had many conversations with Dr. Richard Henderson, one of three scientists awarded the Nobel Prize in Chemistry last week.
“He spent day and night in the lab working on method development for cryo-EM. One of his great contributions was the development of a new type of camera to take better pictures of the proteins studied with cryo-EM,” Dr. Bai said. “When I first moved to England, he gave me a lot of guidance and support and we had many discussions about cryo-EM techniques.”
Co-lead author of the study is Dr. Ji She, a postdoctoral researcher in Physiology and Biophysics. Other contributing authors are Dr. Jiangtao Guo, a postdoctoral fellow in Physiology and Biophysics, research specialist Dr. Weizhong Zeng, and a researcher from the University of Michigan.
In a related Nature study, newly recruited UT Southwestern faculty member Dr. Xiaochun Li, Assistant Professor of Molecular Genetics and Biophysics, determined the structure of the human TRPML1 in the open and closed conformations (positions) in response to a synthetic molecule that opened the channel. That work, also reported online today, was done when Dr. Li was a postdoctoral researcher at Rockefeller University.
Funding: The study received support from the National Institutes of Health, The Welch Foundation, HHMI, and CPRIT.
Source: Deborah Wormser – UT Southwestern
Image Source: NeuroscienceNews.com images is credited to the researchers.
Original Research: Abstract for “Structure of mammalian endolysosomal TRPML1 channel in nanodiscs” by Qingfeng Chen, Ji She, Weizhong Zeng, Jiangtao Guo, Haoxing Xu, Xiao-chen Bai & Youxing Jiang in Nature. Published online October 11 2017 doi:10.1038/nature24035
[cbtabs][cbtab title=”MLA”]UT Southwestern “3D Atomic Structure of Ion Channel Impliacted in Rare Neurodegenerative Disease.” NeuroscienceNews. NeuroscienceNews, 12 October 2017.
<https://neurosciencenews.com/ion-channel-3d-neurodegeneration-7738/>.[/cbtab][cbtab title=”APA”]UT Southwestern (2017, October 12). 3D Atomic Structure of Ion Channel Impliacted in Rare Neurodegenerative Disease. NeuroscienceNews. Retrieved October 12, 2017 from https://neurosciencenews.com/ion-channel-3d-neurodegeneration-7738/[/cbtab][cbtab title=”Chicago”]UT Southwestern “3D Atomic Structure of Ion Channel Impliacted in Rare Neurodegenerative Disease.” https://neurosciencenews.com/ion-channel-3d-neurodegeneration-7738/ (accessed October 12, 2017).[/cbtab][/cbtabs]
Abstract
Structure of mammalian endolysosomal TRPML1 channel in nanodiscs
Transient receptor potential mucolipin 1 (TRPML1) is a cation channel located within endosomal and lysosomal membranes. Ubiquitously expressed in mammalian cells its loss-of-function mutations are the direct cause of type IV mucolipidosis, an autosomal recessive lysosomal storage disease. Here we present the single-particle electron cryo-microscopy structure of the mouse TRPML1 channel embedded in nanodiscs. Combined with mutagenesis analysis, the TRPML1 structure reveals that phosphatidylinositol-3,5-bisphosphate (PtdIns(3,5)P2) binds to the N terminus of the channel—distal from the pore—and the helix–turn–helix extension between segments S2 and S3 probably couples ligand binding to pore opening. The tightly packed selectivity filter contains multiple ion-binding sites, and the conserved acidic residues form the luminal Ca2+-blocking site that confers luminal pH and Ca2+ modulation on channel conductance. A luminal linker domain forms a fenestrated canopy atop the channel, providing several luminal ion passages to the pore and creating a negative electrostatic trap, with a preference for divalent cations, at the luminal entrance. The structure also reveals two equally distributed S4–S5 linker conformations in the closed channel, suggesting an S4–S5 linker-mediated PtdInsP2 gating mechanism among TRPML channels.
“Structure of mammalian endolysosomal TRPML1 channel in nanodiscs” by Qingfeng Chen, Ji She, Weizhong Zeng, Jiangtao Guo, Haoxing Xu, Xiao-chen Bai & Youxing Jiang in Nature. Published online October 11 2017 doi:10.1038/nature24035