Summary: Researchers have successfully reconstructed a short movie of randomly moving discs produced by rat retinal neurons, with the help of machine learning technology.
Source: PLOS.
Using machine-learning techniques, a research team has reconstructed a short movie of small, randomly moving discs from signals produced by rat retinal neurons. Vicente Botella-Soler of the Institute of Science and Technology Austria and colleagues present this work in PLOS Computational Biology.
Neurons in the mammalian retina transform light patterns into electrical signals that are transmitted to the brain. Reconstructing light patterns from neuron signals, a process known as decoding, can help reveal what kind of information these signals carry. However, most decoding efforts to date have used simple stimuli and have relied on small numbers (fewer than 50) of retinal neurons.
In the new study, Botella-Soler and colleagues examined a small patch of about 100 neurons taken from the retina of a rat. They recorded the electrical signals produced by each neuron in response to short movies of small discs moving in a complex, random pattern. The researchers used various regression methods to compare their ability to reconstruct a movie one frame at a time, pixel by pixel.
The research team found that a mathematically simple linear decoder produced an accurate reconstruction of the movie. However, nonlinear methods reconstructed the movie more accurately, and two very different nonlinear methods, neural nets and kernelized decoders, performed similarly well.
Unlike linear decoders, the researchers demonstrated that nonlinear methods were sensitive to each neuron signal in the context of previous signals from the same neuron. The researchers hypothesized that this history dependence enabled the nonlinear decoders to ignore spontaneous neuron signals that do not correspond to an actual stimulus, while a linear decoder might “hallucinate” stimuli in response to such spontaneously generated neural activity.
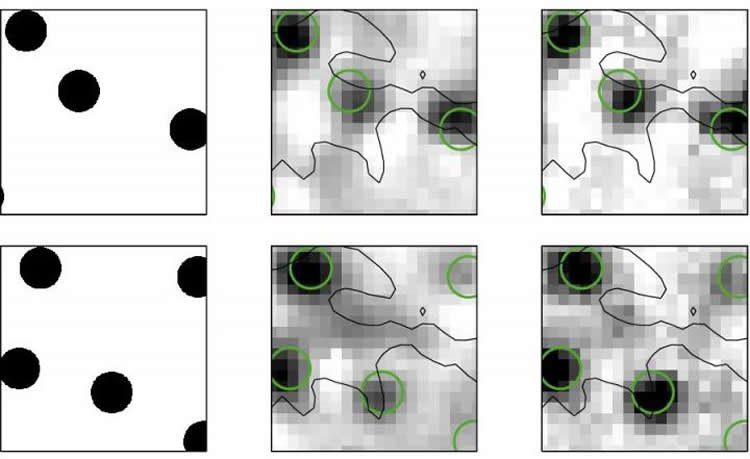
These findings could pave the way to improved decoding methods and better understanding of what different types of retinal neurons do and why they are needed. As a next step, Botella-Soler and colleagues will investigate how well decoders trained on a new class of synthetic stimuli might generalize to both simpler as well as naturally complex stimuli.
“I hope that our work showcases that with sufficient attention to experimental design and computational exploration, it is possible to open the box of modern statistical and machine learning methods and actually interpret which features in the data give rise to their extra predictive power,” says study senior author Gasper Tkacik. “This is the path to not only reporting better quantitative performance, but also extracting new insights and testable hypotheses about biological systems.”
Source: Gasper Tkacik – PLOS
Publisher: Organized by NeuroscienceNews.com.
Image Source: NeuroscienceNews.com image is credited to Botella-Soler et al.
Original Research: Open access research for “Nonlinear decoding of a complex movie from the mammalian retina” by Vicente Botella-Soler, Stéphane Deny, Georg Martius, Olivier Marre, and Gašper Tkačik in PLOS Computational Biology. Published May 10 2018.
doi:10.1371/journal.pcbi.1006057
[cbtabs][cbtab title=”MLA”]PLOS”Video of Moving Discs Reconstructed From Rat Retinal Neural Signals.” NeuroscienceNews. NeuroscienceNews, 11 May 2018.
<https://neurosciencenews.com/disc-movement-rat-retina-9029/>.[/cbtab][cbtab title=”APA”]PLOS(2018, May 11). Video of Moving Discs Reconstructed From Rat Retinal Neural Signals. NeuroscienceNews. Retrieved May 11, 2018 from https://neurosciencenews.com/disc-movement-rat-retina-9029/[/cbtab][cbtab title=”Chicago”]PLOS”Video of Moving Discs Reconstructed From Rat Retinal Neural Signals.” https://neurosciencenews.com/disc-movement-rat-retina-9029/ (accessed May 11, 2018).[/cbtab][/cbtabs]
Abstract
Nonlinear decoding of a complex movie from the mammalian retina
Retina is a paradigmatic system for studying sensory encoding: the transformation of light into spiking activity of ganglion cells. The inverse problem, where stimulus is reconstructed from spikes, has received less attention, especially for complex stimuli that should be reconstructed “pixel-by-pixel”. We recorded around a hundred neurons from a dense patch in a rat retina and decoded movies of multiple small randomly-moving discs. We constructed nonlinear (kernelized and neural network) decoders that improved significantly over linear results. An important contribution to this was the ability of nonlinear decoders to reliably separate between neural responses driven by locally fluctuating light signals, and responses at locally constant light driven by spontaneous-like activity. This improvement crucially depended on the precise, non-Poisson temporal structure of individual spike trains, which originated in the spike-history dependence of neural responses. We propose a general principle by which downstream circuitry could discriminate between spontaneous and stimulus-driven activity based solely on higher-order statistical structure in the incoming spike trains.